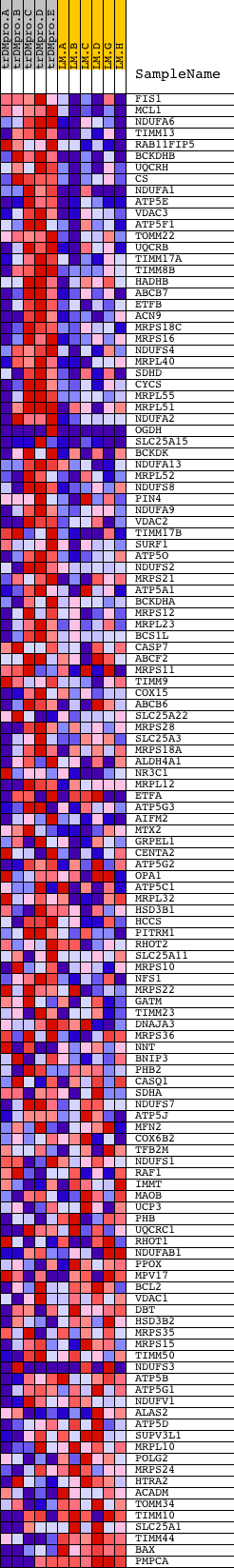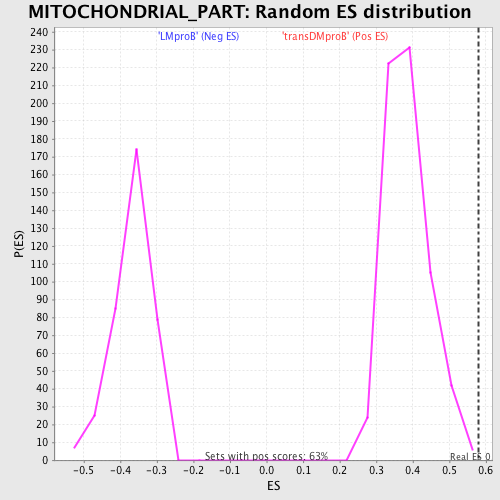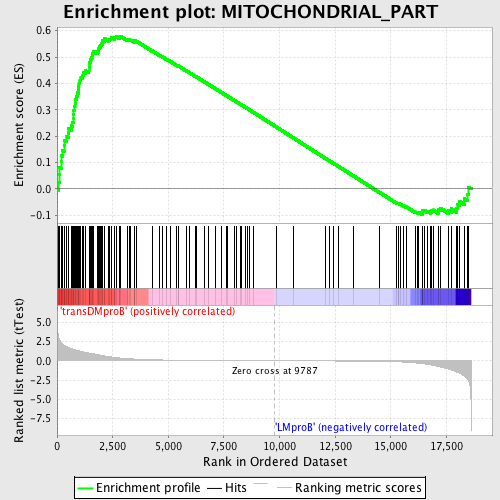
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | MITOCHONDRIAL_PART |
| Enrichment Score (ES) | 0.579467 |
| Normalized Enrichment Score (NES) | 1.5043613 |
| Nominal p-value | 0.0015873016 |
| FDR q-value | 0.48791066 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FIS1 | 16669 3579 | 81 | 2.887 | 0.0263 | Yes | ||
| 2 | MCL1 | 15502 | 99 | 2.742 | 0.0545 | Yes | ||
| 3 | NDUFA6 | 12473 | 107 | 2.667 | 0.0825 | Yes | ||
| 4 | TIMM13 | 14974 | 179 | 2.310 | 0.1032 | Yes | ||
| 5 | RAB11FIP5 | 17089 | 187 | 2.296 | 0.1272 | Yes | ||
| 6 | BCKDHB | 19365 | 242 | 2.161 | 0.1473 | Yes | ||
| 7 | UQCRH | 12378 | 330 | 1.989 | 0.1637 | Yes | ||
| 8 | CS | 19839 | 345 | 1.955 | 0.1837 | Yes | ||
| 9 | NDUFA1 | 24170 | 427 | 1.811 | 0.1986 | Yes | ||
| 10 | ATP5E | 14321 | 506 | 1.707 | 0.2125 | Yes | ||
| 11 | VDAC3 | 10284 | 520 | 1.693 | 0.2298 | Yes | ||
| 12 | ATP5F1 | 15212 | 656 | 1.576 | 0.2392 | Yes | ||
| 13 | TOMM22 | 5863 | 689 | 1.540 | 0.2539 | Yes | ||
| 14 | UQCRB | 12545 | 735 | 1.494 | 0.2673 | Yes | ||
| 15 | TIMM17A | 13822 | 739 | 1.490 | 0.2830 | Yes | ||
| 16 | TIMM8B | 11391 | 749 | 1.481 | 0.2982 | Yes | ||
| 17 | HADHB | 3514 10527 | 764 | 1.463 | 0.3130 | Yes | ||
| 18 | ABCB7 | 24085 | 805 | 1.427 | 0.3260 | Yes | ||
| 19 | ETFB | 18279 | 837 | 1.400 | 0.3392 | Yes | ||
| 20 | ACN9 | 12920 | 873 | 1.366 | 0.3518 | Yes | ||
| 21 | MRPS18C | 12728 | 898 | 1.345 | 0.3648 | Yes | ||
| 22 | MRPS16 | 12311 | 953 | 1.301 | 0.3758 | Yes | ||
| 23 | NDUFS4 | 9452 5157 3173 | 969 | 1.290 | 0.3887 | Yes | ||
| 24 | MRPL40 | 22641 | 982 | 1.285 | 0.4017 | Yes | ||
| 25 | SDHD | 19125 | 1026 | 1.248 | 0.4126 | Yes | ||
| 26 | CYCS | 8821 | 1071 | 1.216 | 0.4231 | Yes | ||
| 27 | MRPL55 | 20864 | 1157 | 1.163 | 0.4309 | Yes | ||
| 28 | MRPL51 | 12361 7301 | 1184 | 1.141 | 0.4416 | Yes | ||
| 29 | NDUFA2 | 9450 | 1265 | 1.089 | 0.4489 | Yes | ||
| 30 | OGDH | 9502 1412 | 1433 | 0.996 | 0.4504 | Yes | ||
| 31 | SLC25A15 | 356 3819 | 1436 | 0.995 | 0.4609 | Yes | ||
| 32 | BCKDK | 18061 1391 | 1447 | 0.990 | 0.4709 | Yes | ||
| 33 | NDUFA13 | 7400 | 1451 | 0.990 | 0.4812 | Yes | ||
| 34 | MRPL52 | 22019 | 1492 | 0.974 | 0.4894 | Yes | ||
| 35 | NDUFS8 | 23950 | 1524 | 0.958 | 0.4979 | Yes | ||
| 36 | PIN4 | 12817 | 1569 | 0.934 | 0.5055 | Yes | ||
| 37 | NDUFA9 | 12288 | 1595 | 0.923 | 0.5139 | Yes | ||
| 38 | VDAC2 | 22081 | 1629 | 0.901 | 0.5217 | Yes | ||
| 39 | TIMM17B | 24390 | 1806 | 0.797 | 0.5207 | Yes | ||
| 40 | SURF1 | 14645 | 1872 | 0.761 | 0.5252 | Yes | ||
| 41 | ATP5O | 22539 | 1878 | 0.758 | 0.5330 | Yes | ||
| 42 | NDUFS2 | 4089 13760 | 1905 | 0.742 | 0.5395 | Yes | ||
| 43 | MRPS21 | 12323 | 1960 | 0.713 | 0.5442 | Yes | ||
| 44 | ATP5A1 | 23505 | 1993 | 0.692 | 0.5498 | Yes | ||
| 45 | BCKDHA | 17925 | 2027 | 0.678 | 0.5552 | Yes | ||
| 46 | MRPS12 | 17907 6273 | 2036 | 0.672 | 0.5619 | Yes | ||
| 47 | MRPL23 | 9738 | 2125 | 0.630 | 0.5639 | Yes | ||
| 48 | BCS1L | 14223 | 2140 | 0.622 | 0.5697 | Yes | ||
| 49 | CASP7 | 23826 | 2307 | 0.540 | 0.5665 | Yes | ||
| 50 | ABCF2 | 11338 6580 6579 | 2376 | 0.512 | 0.5682 | Yes | ||
| 51 | MRPS11 | 12621 3150 | 2424 | 0.497 | 0.5710 | Yes | ||
| 52 | TIMM9 | 19686 | 2450 | 0.488 | 0.5748 | Yes | ||
| 53 | COX15 | 5931 | 2588 | 0.431 | 0.5720 | Yes | ||
| 54 | ABCB6 | 13162 | 2597 | 0.429 | 0.5761 | Yes | ||
| 55 | SLC25A22 | 12671 | 2655 | 0.411 | 0.5774 | Yes | ||
| 56 | MRPS28 | 15377 | 2800 | 0.362 | 0.5734 | Yes | ||
| 57 | SLC25A3 | 19644 | 2818 | 0.359 | 0.5763 | Yes | ||
| 58 | MRPS18A | 23222 | 2831 | 0.355 | 0.5795 | Yes | ||
| 59 | ALDH4A1 | 16011 | 3152 | 0.277 | 0.5651 | No | ||
| 60 | NR3C1 | 9043 | 3177 | 0.270 | 0.5667 | No | ||
| 61 | MRPL12 | 12090 | 3252 | 0.256 | 0.5654 | No | ||
| 62 | ETFA | 4292 8495 3155 | 3312 | 0.244 | 0.5648 | No | ||
| 63 | ATP5G3 | 14558 | 3463 | 0.219 | 0.5590 | No | ||
| 64 | AIFM2 | 3320 3407 20006 3317 | 3478 | 0.218 | 0.5606 | No | ||
| 65 | MTX2 | 14980 | 3489 | 0.215 | 0.5623 | No | ||
| 66 | GRPEL1 | 16863 | 3556 | 0.205 | 0.5609 | No | ||
| 67 | CENTA2 | 20748 | 4276 | 0.126 | 0.5234 | No | ||
| 68 | ATP5G2 | 12610 | 4623 | 0.105 | 0.5058 | No | ||
| 69 | OPA1 | 13169 22800 7894 | 4726 | 0.099 | 0.5013 | No | ||
| 70 | ATP5C1 | 8635 | 4914 | 0.089 | 0.4921 | No | ||
| 71 | MRPL32 | 7961 | 5111 | 0.081 | 0.4824 | No | ||
| 72 | HSD3B1 | 15227 | 5362 | 0.072 | 0.4696 | No | ||
| 73 | HCCS | 9079 4842 | 5438 | 0.070 | 0.4663 | No | ||
| 74 | PITRM1 | 21710 | 5442 | 0.070 | 0.4669 | No | ||
| 75 | RHOT2 | 1533 10073 | 5836 | 0.058 | 0.4463 | No | ||
| 76 | SLC25A11 | 20366 1368 | 5952 | 0.056 | 0.4406 | No | ||
| 77 | MRPS10 | 7229 12261 23212 | 6204 | 0.049 | 0.4276 | No | ||
| 78 | NFS1 | 2903 14377 | 6281 | 0.047 | 0.4240 | No | ||
| 79 | MRPS22 | 19031 | 6626 | 0.041 | 0.4058 | No | ||
| 80 | GATM | 14451 | 6815 | 0.037 | 0.3960 | No | ||
| 81 | TIMM23 | 7004 5768 | 7129 | 0.032 | 0.3794 | No | ||
| 82 | DNAJA3 | 1732 13518 | 7396 | 0.028 | 0.3654 | No | ||
| 83 | MRPS36 | 7257 | 7612 | 0.025 | 0.3540 | No | ||
| 84 | NNT | 3273 5181 9471 3238 | 7663 | 0.024 | 0.3516 | No | ||
| 85 | BNIP3 | 17581 | 7980 | 0.020 | 0.3347 | No | ||
| 86 | PHB2 | 17286 | 8063 | 0.019 | 0.3304 | No | ||
| 87 | CASQ1 | 13752 8695 | 8234 | 0.017 | 0.3214 | No | ||
| 88 | SDHA | 12436 | 8271 | 0.017 | 0.3197 | No | ||
| 89 | NDUFS7 | 19946 | 8476 | 0.014 | 0.3088 | No | ||
| 90 | ATP5J | 880 22555 | 8543 | 0.014 | 0.3053 | No | ||
| 91 | MFN2 | 15678 2417 | 8669 | 0.012 | 0.2987 | No | ||
| 92 | COX6B2 | 17983 4110 | 8836 | 0.010 | 0.2898 | No | ||
| 93 | TFB2M | 4024 9092 4083 | 9873 | -0.001 | 0.2338 | No | ||
| 94 | NDUFS1 | 13940 | 10640 | -0.009 | 0.1925 | No | ||
| 95 | RAF1 | 17035 | 12066 | -0.026 | 0.1157 | No | ||
| 96 | IMMT | 367 8029 | 12249 | -0.029 | 0.1061 | No | ||
| 97 | MAOB | 24182 2556 | 12439 | -0.031 | 0.0962 | No | ||
| 98 | UCP3 | 3972 18175 | 12653 | -0.035 | 0.0851 | No | ||
| 99 | PHB | 9558 | 13331 | -0.047 | 0.0490 | No | ||
| 100 | UQCRC1 | 10259 | 14490 | -0.081 | -0.0128 | No | ||
| 101 | RHOT1 | 12227 | 15265 | -0.132 | -0.0533 | No | ||
| 102 | NDUFAB1 | 7667 | 15329 | -0.139 | -0.0552 | No | ||
| 103 | PPOX | 13761 4104 | 15352 | -0.141 | -0.0549 | No | ||
| 104 | MPV17 | 9407 3505 3543 | 15441 | -0.152 | -0.0580 | No | ||
| 105 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 15568 | -0.167 | -0.0631 | No | ||
| 106 | VDAC1 | 10282 | 15698 | -0.188 | -0.0680 | No | ||
| 107 | DBT | 1770 4599 | 16108 | -0.279 | -0.0872 | No | ||
| 108 | HSD3B2 | 4873 4874 15229 4875 9126 | 16196 | -0.304 | -0.0887 | No | ||
| 109 | MRPS35 | 17237 1068 | 16245 | -0.318 | -0.0879 | No | ||
| 110 | MRPS15 | 2442 12345 | 16409 | -0.374 | -0.0927 | No | ||
| 111 | TIMM50 | 17911 | 16420 | -0.377 | -0.0893 | No | ||
| 112 | NDUFS3 | 7553 12683 | 16422 | -0.378 | -0.0853 | No | ||
| 113 | ATP5B | 19846 | 16431 | -0.383 | -0.0817 | No | ||
| 114 | ATP5G1 | 8636 | 16499 | -0.407 | -0.0810 | No | ||
| 115 | NDUFV1 | 23953 | 16637 | -0.452 | -0.0836 | No | ||
| 116 | ALAS2 | 24229 | 16801 | -0.536 | -0.0867 | No | ||
| 117 | ATP5D | 19949 | 16843 | -0.556 | -0.0830 | No | ||
| 118 | SUPV3L1 | 11677 | 16897 | -0.585 | -0.0797 | No | ||
| 119 | MRPL10 | 20681 | 17128 | -0.725 | -0.0844 | No | ||
| 120 | POLG2 | 20180 | 17158 | -0.748 | -0.0780 | No | ||
| 121 | MRPS24 | 12262 | 17238 | -0.820 | -0.0736 | No | ||
| 122 | HTRA2 | 1122 17103 | 17598 | -1.073 | -0.0816 | No | ||
| 123 | ACADM | 15133 1800 | 17712 | -1.192 | -0.0750 | No | ||
| 124 | TOMM34 | 14359 | 17955 | -1.452 | -0.0727 | No | ||
| 125 | TIMM10 | 11390 2158 | 17991 | -1.495 | -0.0587 | No | ||
| 126 | SLC25A1 | 22645 | 18089 | -1.611 | -0.0468 | No | ||
| 127 | TIMM44 | 3858 18941 | 18303 | -2.004 | -0.0370 | No | ||
| 128 | BAX | 17832 | 18451 | -2.419 | -0.0193 | No | ||
| 129 | PMPCA | 12421 7350 | 18502 | -2.649 | 0.0062 | No |