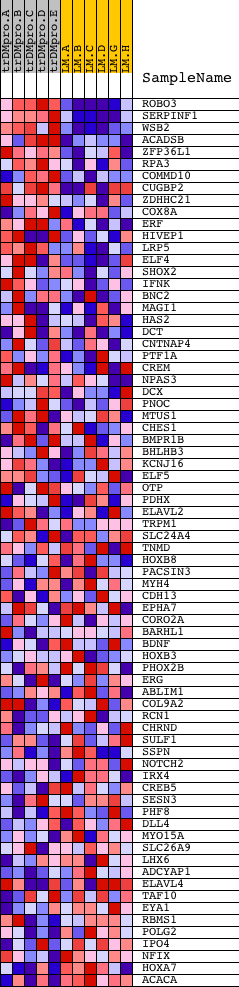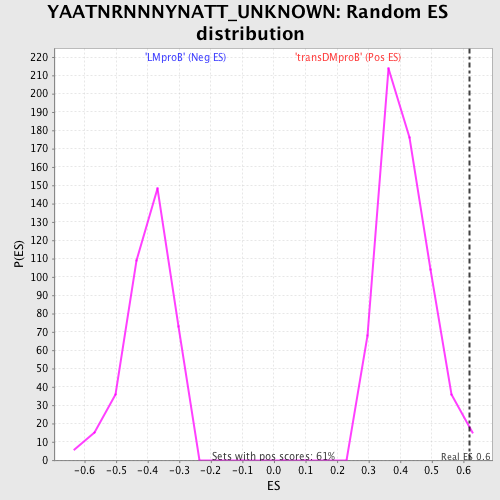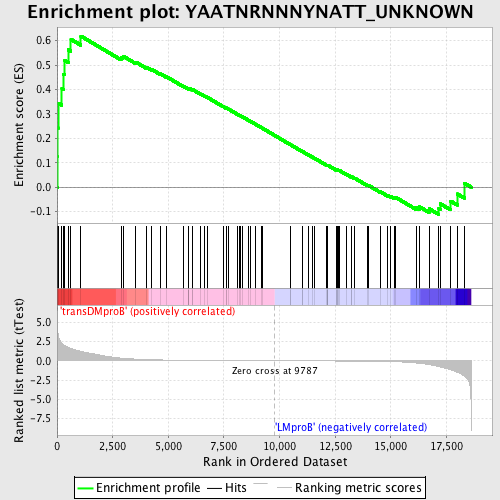
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | YAATNRNNNYNATT_UNKNOWN |
| Enrichment Score (ES) | 0.6186096 |
| Normalized Enrichment Score (NES) | 1.4886396 |
| Nominal p-value | 0.0146818925 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.904 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ROBO3 | 9705 | 18 | 4.175 | 0.1263 | Yes | ||
| 2 | SERPINF1 | 9793 | 25 | 3.846 | 0.2432 | Yes | ||
| 3 | WSB2 | 7205 | 45 | 3.364 | 0.3448 | Yes | ||
| 4 | ACADSB | 7358 | 216 | 2.222 | 0.4034 | Yes | ||
| 5 | ZFP36L1 | 4453 4454 | 276 | 2.073 | 0.4634 | Yes | ||
| 6 | RPA3 | 12667 | 346 | 1.954 | 0.5192 | Yes | ||
| 7 | COMMD10 | 23561 | 507 | 1.706 | 0.5626 | Yes | ||
| 8 | CUGBP2 | 4687 8923 | 623 | 1.608 | 0.6054 | Yes | ||
| 9 | ZDHHC21 | 15854 | 1069 | 1.220 | 0.6186 | Yes | ||
| 10 | COX8A | 8777 | 2879 | 0.344 | 0.5315 | No | ||
| 11 | ERF | 4680 8914 | 2962 | 0.320 | 0.5369 | No | ||
| 12 | HIVEP1 | 8486 | 3531 | 0.209 | 0.5126 | No | ||
| 13 | LRP5 | 23948 9285 | 4008 | 0.151 | 0.4915 | No | ||
| 14 | ELF4 | 24162 | 4264 | 0.127 | 0.4816 | No | ||
| 15 | SHOX2 | 1802 5432 | 4632 | 0.104 | 0.4650 | No | ||
| 16 | IFNK | 11837 | 4924 | 0.089 | 0.4520 | No | ||
| 17 | BNC2 | 15850 | 5676 | 0.063 | 0.4134 | No | ||
| 18 | MAGI1 | 19828 4816 1179 | 5904 | 0.057 | 0.4029 | No | ||
| 19 | HAS2 | 22293 | 5907 | 0.057 | 0.4045 | No | ||
| 20 | DCT | 4603 21723 | 5920 | 0.056 | 0.4056 | No | ||
| 21 | CNTNAP4 | 18462 3857 | 6063 | 0.053 | 0.3996 | No | ||
| 22 | PTF1A | 15110 | 6087 | 0.052 | 0.3999 | No | ||
| 23 | CREM | 2005 1965 23496 1951 | 6453 | 0.044 | 0.3816 | No | ||
| 24 | NPAS3 | 2077 21273 | 6641 | 0.041 | 0.3727 | No | ||
| 25 | DCX | 24040 8841 4604 | 6742 | 0.039 | 0.3685 | No | ||
| 26 | PNOC | 21783 | 7478 | 0.027 | 0.3297 | No | ||
| 27 | MTUS1 | 4162 158 | 7595 | 0.025 | 0.3242 | No | ||
| 28 | CHES1 | 21007 11200 | 7601 | 0.025 | 0.3247 | No | ||
| 29 | BMPR1B | 8661 15152 4451 | 7723 | 0.023 | 0.3189 | No | ||
| 30 | BHLHB3 | 16935 | 8115 | 0.018 | 0.2984 | No | ||
| 31 | KCNJ16 | 20613 | 8218 | 0.017 | 0.2934 | No | ||
| 32 | ELF5 | 14936 2913 | 8251 | 0.017 | 0.2922 | No | ||
| 33 | OTP | 21582 | 8333 | 0.016 | 0.2883 | No | ||
| 34 | PDHX | 14510 | 8608 | 0.013 | 0.2739 | No | ||
| 35 | ELAVL2 | 15839 | 8685 | 0.012 | 0.2702 | No | ||
| 36 | TRPM1 | 10846 474 9395 494 | 8933 | 0.009 | 0.2571 | No | ||
| 37 | SLC24A4 | 6223 | 9190 | 0.006 | 0.2435 | No | ||
| 38 | TNMD | 24260 | 9252 | 0.006 | 0.2404 | No | ||
| 39 | HOXB8 | 9111 | 10477 | -0.008 | 0.1746 | No | ||
| 40 | PACSIN3 | 14948 8149 2776 2665 | 10478 | -0.008 | 0.1748 | No | ||
| 41 | MYH4 | 20837 | 11042 | -0.014 | 0.1449 | No | ||
| 42 | CDH13 | 4506 3826 | 11044 | -0.014 | 0.1453 | No | ||
| 43 | EPHA7 | 8909 4674 | 11300 | -0.016 | 0.1320 | No | ||
| 44 | CORO2A | 4216 8411 | 11460 | -0.018 | 0.1240 | No | ||
| 45 | BARHL1 | 14634 | 11586 | -0.020 | 0.1179 | No | ||
| 46 | BDNF | 14926 2797 | 12118 | -0.027 | 0.0900 | No | ||
| 47 | HOXB3 | 20685 | 12141 | -0.027 | 0.0897 | No | ||
| 48 | PHOX2B | 16524 | 12143 | -0.027 | 0.0904 | No | ||
| 49 | ERG | 1686 8915 | 12169 | -0.027 | 0.0899 | No | ||
| 50 | ABLIM1 | 3717 3766 3679 | 12548 | -0.033 | 0.0705 | No | ||
| 51 | COL9A2 | 8770 | 12569 | -0.033 | 0.0705 | No | ||
| 52 | RCN1 | 9710 | 12589 | -0.033 | 0.0705 | No | ||
| 53 | CHRND | 3931 4084 | 12591 | -0.033 | 0.0714 | No | ||
| 54 | SULF1 | 6285 | 12630 | -0.034 | 0.0704 | No | ||
| 55 | SSPN | 9246 | 12679 | -0.035 | 0.0689 | No | ||
| 56 | NOTCH2 | 15485 | 13013 | -0.041 | 0.0522 | No | ||
| 57 | IRX4 | 11944 | 13212 | -0.045 | 0.0429 | No | ||
| 58 | CREB5 | 10551 | 13249 | -0.045 | 0.0423 | No | ||
| 59 | SESN3 | 19561 | 13384 | -0.048 | 0.0366 | No | ||
| 60 | PHF8 | 6770 | 13956 | -0.063 | 0.0077 | No | ||
| 61 | DLL4 | 7040 | 14005 | -0.064 | 0.0071 | No | ||
| 62 | MYO15A | 1242 20860 | 14514 | -0.082 | -0.0178 | No | ||
| 63 | SLC26A9 | 11560 980 4067 | 14868 | -0.101 | -0.0338 | No | ||
| 64 | LHX6 | 9275 2914 | 14991 | -0.110 | -0.0370 | No | ||
| 65 | ADCYAP1 | 4348 | 15180 | -0.125 | -0.0434 | No | ||
| 66 | ELAVL4 | 15805 4889 9137 | 15206 | -0.128 | -0.0408 | No | ||
| 67 | TAF10 | 10785 | 16157 | -0.294 | -0.0831 | No | ||
| 68 | EYA1 | 4695 4061 | 16293 | -0.331 | -0.0803 | No | ||
| 69 | RBMS1 | 14580 | 16721 | -0.499 | -0.0881 | No | ||
| 70 | POLG2 | 20180 | 17158 | -0.748 | -0.0888 | No | ||
| 71 | IPO4 | 7983 | 17221 | -0.804 | -0.0677 | No | ||
| 72 | NFIX | 9458 | 17695 | -1.179 | -0.0572 | No | ||
| 73 | HOXA7 | 4862 | 17990 | -1.493 | -0.0276 | No | ||
| 74 | ACACA | 309 4209 | 18306 | -2.010 | 0.0167 | No |