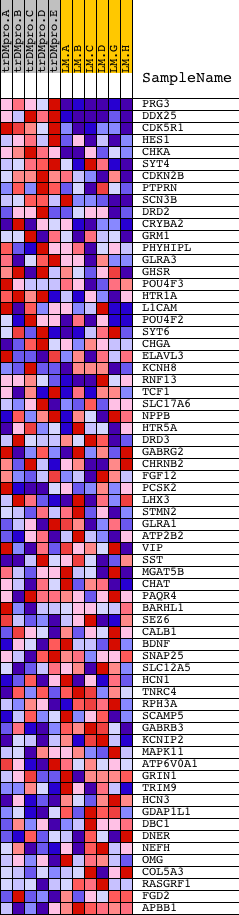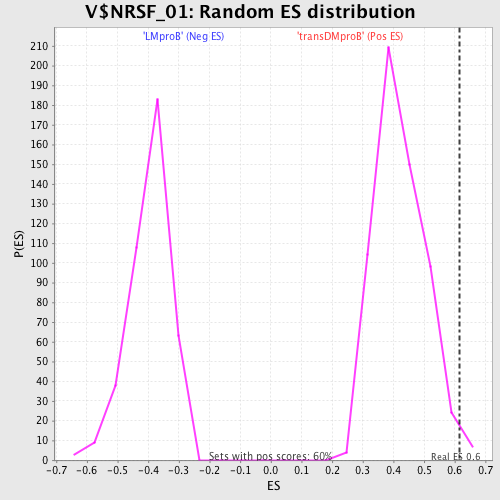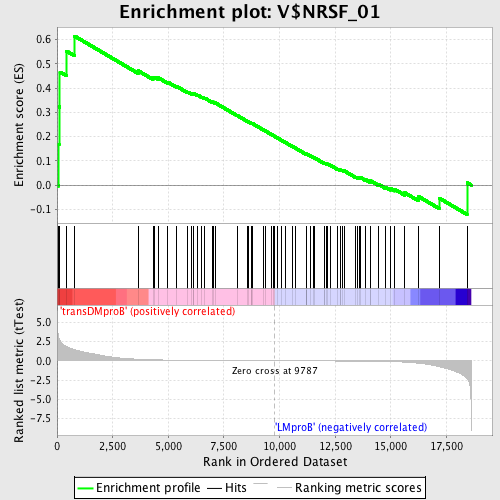
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | V$NRSF_01 |
| Enrichment Score (ES) | 0.6147032 |
| Normalized Enrichment Score (NES) | 1.4584048 |
| Nominal p-value | 0.013422819 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.966 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PRG3 | 14971 | 65 | 3.022 | 0.1671 | Yes | ||
| 2 | DDX25 | 19184 | 90 | 2.792 | 0.3234 | Yes | ||
| 3 | CDK5R1 | 20745 | 128 | 2.574 | 0.4667 | Yes | ||
| 4 | HES1 | 22798 | 430 | 1.809 | 0.5526 | Yes | ||
| 5 | CHKA | 23759 | 789 | 1.442 | 0.6147 | Yes | ||
| 6 | SYT4 | 5566 | 3643 | 0.193 | 0.4718 | No | ||
| 7 | CDKN2B | 15840 | 4312 | 0.123 | 0.4427 | No | ||
| 8 | PTPRN | 13914 | 4384 | 0.118 | 0.4456 | No | ||
| 9 | SCN3B | 39 3117 6163 10646 | 4547 | 0.109 | 0.4430 | No | ||
| 10 | DRD2 | 19461 | 4971 | 0.087 | 0.4250 | No | ||
| 11 | CRYBA2 | 13920 | 5387 | 0.072 | 0.4067 | No | ||
| 12 | GRM1 | 19822 | 5878 | 0.057 | 0.3835 | No | ||
| 13 | PHYHIPL | 12907 7711 | 6046 | 0.053 | 0.3775 | No | ||
| 14 | GLRA3 | 18617 | 6108 | 0.052 | 0.3771 | No | ||
| 15 | GHSR | 1810 15625 | 6123 | 0.051 | 0.3793 | No | ||
| 16 | POU4F3 | 9604 | 6323 | 0.046 | 0.3712 | No | ||
| 17 | HTR1A | 21569 | 6510 | 0.043 | 0.3636 | No | ||
| 18 | L1CAM | 3 | 6639 | 0.041 | 0.3590 | No | ||
| 19 | POU4F2 | 9603 5276 | 7003 | 0.034 | 0.3413 | No | ||
| 20 | SYT6 | 13437 7042 12040 | 7007 | 0.034 | 0.3431 | No | ||
| 21 | CHGA | 21178 2151 | 7102 | 0.033 | 0.3399 | No | ||
| 22 | ELAVL3 | 9136 | 8116 | 0.018 | 0.2863 | No | ||
| 23 | KCNH8 | 23197 | 8562 | 0.013 | 0.2631 | No | ||
| 24 | RNF13 | 6271 | 8619 | 0.013 | 0.2608 | No | ||
| 25 | TCF1 | 16416 | 8723 | 0.011 | 0.2559 | No | ||
| 26 | SLC17A6 | 18226 | 8756 | 0.011 | 0.2548 | No | ||
| 27 | NPPB | 15994 | 8787 | 0.011 | 0.2538 | No | ||
| 28 | HTR5A | 16899 | 9281 | 0.005 | 0.2275 | No | ||
| 29 | DRD3 | 22750 | 9363 | 0.005 | 0.2234 | No | ||
| 30 | GABRG2 | 1259 4748 8996 1331 | 9637 | 0.002 | 0.2088 | No | ||
| 31 | CHRNB2 | 8532 4321 | 9705 | 0.001 | 0.2052 | No | ||
| 32 | FGF12 | 1723 22621 | 9778 | 0.000 | 0.2013 | No | ||
| 33 | PCSK2 | 9537 5231 14824 | 9916 | -0.002 | 0.1940 | No | ||
| 34 | LHX3 | 2761 4996 | 10067 | -0.003 | 0.1861 | No | ||
| 35 | STMN2 | 15638 | 10263 | -0.005 | 0.1759 | No | ||
| 36 | GLRA1 | 9021 | 10567 | -0.008 | 0.1601 | No | ||
| 37 | ATP2B2 | 17038 | 10699 | -0.010 | 0.1535 | No | ||
| 38 | VIP | 20096 | 11191 | -0.015 | 0.1279 | No | ||
| 39 | SST | 22625 | 11194 | -0.015 | 0.1287 | No | ||
| 40 | MGAT5B | 20584 11197 | 11209 | -0.015 | 0.1288 | No | ||
| 41 | CHAT | 21882 | 11375 | -0.017 | 0.1209 | No | ||
| 42 | PAQR4 | 23105 | 11508 | -0.019 | 0.1148 | No | ||
| 43 | BARHL1 | 14634 | 11586 | -0.020 | 0.1118 | No | ||
| 44 | SEZ6 | 20764 | 11999 | -0.025 | 0.0910 | No | ||
| 45 | CALB1 | 4469 | 12020 | -0.026 | 0.0914 | No | ||
| 46 | BDNF | 14926 2797 | 12118 | -0.027 | 0.0877 | No | ||
| 47 | SNAP25 | 5466 | 12150 | -0.027 | 0.0875 | No | ||
| 48 | SLC12A5 | 14731 | 12288 | -0.029 | 0.0818 | No | ||
| 49 | HCN1 | 21557 | 12616 | -0.034 | 0.0661 | No | ||
| 50 | TNRC4 | 15514 | 12751 | -0.036 | 0.0609 | No | ||
| 51 | RPH3A | 13628 | 12753 | -0.036 | 0.0629 | No | ||
| 52 | SCAMP5 | 3028 19100 | 12822 | -0.037 | 0.0613 | No | ||
| 53 | GABRB3 | 4747 | 12896 | -0.039 | 0.0596 | No | ||
| 54 | KCNIP2 | 23655 3725 | 13428 | -0.049 | 0.0337 | No | ||
| 55 | MAPK11 | 2264 9618 22163 | 13520 | -0.051 | 0.0317 | No | ||
| 56 | ATP6V0A1 | 8640 4424 1197 | 13586 | -0.053 | 0.0312 | No | ||
| 57 | GRIN1 | 9041 4804 | 13657 | -0.055 | 0.0305 | No | ||
| 58 | TRIM9 | 2118 8076 13577 8234 | 13851 | -0.060 | 0.0235 | No | ||
| 59 | HCN3 | 4843 1763 | 14085 | -0.067 | 0.0147 | No | ||
| 60 | GDAP1L1 | 14747 | 14091 | -0.067 | 0.0182 | No | ||
| 61 | DBC1 | 7147 | 14457 | -0.080 | 0.0030 | No | ||
| 62 | DNER | 13901 | 14764 | -0.094 | -0.0082 | No | ||
| 63 | NEFH | 11724 | 14986 | -0.109 | -0.0139 | No | ||
| 64 | OMG | 5209 | 15174 | -0.125 | -0.0170 | No | ||
| 65 | COL5A3 | 19219 | 15629 | -0.178 | -0.0314 | No | ||
| 66 | RASGRF1 | 3050 19357 | 16256 | -0.320 | -0.0471 | No | ||
| 67 | FGD2 | 23309 | 17181 | -0.766 | -0.0536 | No | ||
| 68 | APBB1 | 17705 | 18429 | -2.320 | 0.0101 | No |