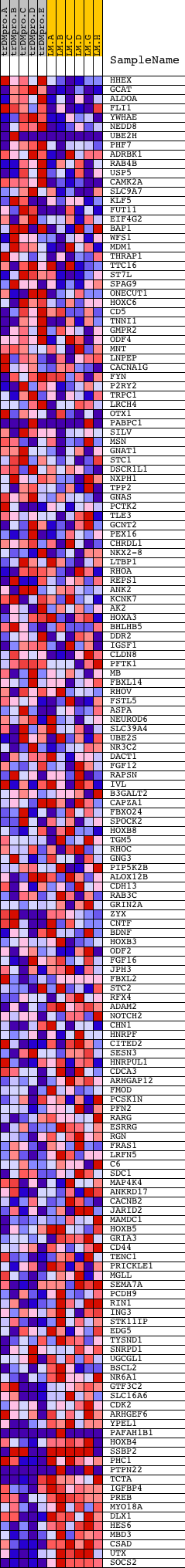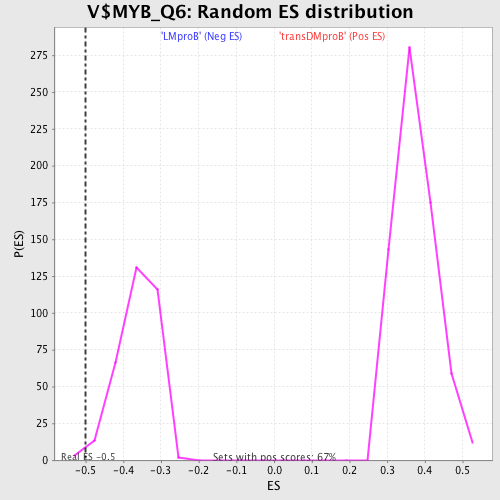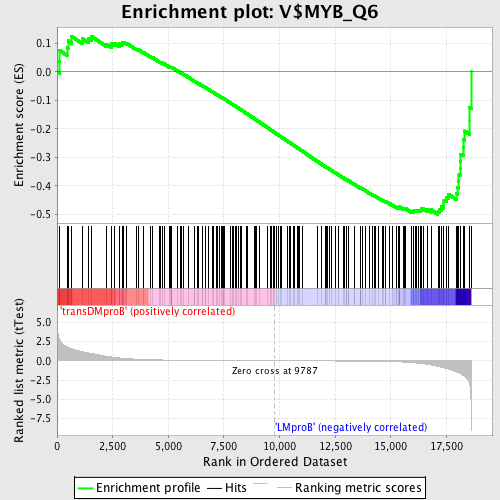
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | V$MYB_Q6 |
| Enrichment Score (ES) | -0.50024563 |
| Normalized Enrichment Score (NES) | -1.3854196 |
| Nominal p-value | 0.009063444 |
| FDR q-value | 0.636743 |
| FWER p-Value | 0.996 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HHEX | 23872 | 102 | 2.715 | 0.0363 | No | ||
| 2 | GCAT | 11234 | 121 | 2.601 | 0.0753 | No | ||
| 3 | ALDOA | 8572 | 447 | 1.786 | 0.0852 | No | ||
| 4 | FLI1 | 4729 | 493 | 1.724 | 0.1092 | No | ||
| 5 | YWHAE | 20776 | 651 | 1.581 | 0.1251 | No | ||
| 6 | NEDD8 | 21822 | 1130 | 1.180 | 0.1173 | No | ||
| 7 | UBE2H | 5823 5822 | 1406 | 1.002 | 0.1178 | No | ||
| 8 | PHF7 | 21890 | 1534 | 0.954 | 0.1256 | No | ||
| 9 | ADRBK1 | 23960 | 2237 | 0.568 | 0.0963 | No | ||
| 10 | RAB4B | 5345 | 2435 | 0.493 | 0.0932 | No | ||
| 11 | USP5 | 17004 | 2459 | 0.484 | 0.0994 | No | ||
| 12 | CAMK2A | 2024 23541 1980 | 2586 | 0.432 | 0.0993 | No | ||
| 13 | SLC9A7 | 24177 | 2796 | 0.362 | 0.0935 | No | ||
| 14 | KLF5 | 4456 | 2802 | 0.361 | 0.0988 | No | ||
| 15 | FUT11 | 13084 | 2920 | 0.333 | 0.0976 | No | ||
| 16 | EIF4G2 | 1908 8892 | 2927 | 0.331 | 0.1024 | No | ||
| 17 | BAP1 | 22057 | 2982 | 0.314 | 0.1042 | No | ||
| 18 | WFS1 | 16548 | 3133 | 0.280 | 0.1004 | No | ||
| 19 | MDM1 | 3306 3370 3316 3405 3376 19870 3429 3398 3406 3428 3346 355 | 3567 | 0.203 | 0.0801 | No | ||
| 20 | THRAP1 | 20309 | 3639 | 0.194 | 0.0792 | No | ||
| 21 | TTC16 | 14618 | 3899 | 0.162 | 0.0677 | No | ||
| 22 | ST7L | 15465 1904 | 4216 | 0.131 | 0.0526 | No | ||
| 23 | SPAG9 | 1322 12904 7707 | 4304 | 0.123 | 0.0497 | No | ||
| 24 | ONECUT1 | 4858 | 4617 | 0.105 | 0.0345 | No | ||
| 25 | HOXC6 | 22339 2302 | 4655 | 0.103 | 0.0340 | No | ||
| 26 | CD5 | 23741 | 4721 | 0.099 | 0.0320 | No | ||
| 27 | TNNI1 | 14116 | 4809 | 0.094 | 0.0288 | No | ||
| 28 | GMPR2 | 2711 | 5041 | 0.084 | 0.0175 | No | ||
| 29 | ODF4 | 20398 | 5092 | 0.082 | 0.0161 | No | ||
| 30 | MNT | 20784 | 5133 | 0.080 | 0.0152 | No | ||
| 31 | LNPEP | 6267 10760 10761 | 5137 | 0.080 | 0.0162 | No | ||
| 32 | CACNA1G | 1237 940 20292 | 5162 | 0.079 | 0.0161 | No | ||
| 33 | FYN | 3375 3395 20052 | 5417 | 0.071 | 0.0035 | No | ||
| 34 | P2RY2 | 17736 1279 | 5562 | 0.066 | -0.0033 | No | ||
| 35 | TRPC1 | 2984 19039 | 5606 | 0.065 | -0.0047 | No | ||
| 36 | LRCH4 | 10544 6090 | 5693 | 0.062 | -0.0084 | No | ||
| 37 | OTX1 | 20517 | 5909 | 0.056 | -0.0191 | No | ||
| 38 | PABPC1 | 5219 9522 9523 23572 | 6169 | 0.050 | -0.0324 | No | ||
| 39 | SILV | 16130 | 6187 | 0.050 | -0.0326 | No | ||
| 40 | MSN | 5122 | 6292 | 0.047 | -0.0375 | No | ||
| 41 | GNAT1 | 3027 19000 | 6353 | 0.046 | -0.0400 | No | ||
| 42 | STC1 | 21974 | 6513 | 0.043 | -0.0480 | No | ||
| 43 | DSCR1L1 | 23231 | 6554 | 0.042 | -0.0495 | No | ||
| 44 | NXPH1 | 17524 | 6679 | 0.040 | -0.0556 | No | ||
| 45 | TPP2 | 14257 | 6688 | 0.040 | -0.0554 | No | ||
| 46 | GNAS | 9025 2963 2752 | 6786 | 0.038 | -0.0601 | No | ||
| 47 | PCTK2 | 19907 | 6966 | 0.035 | -0.0693 | No | ||
| 48 | TLE3 | 5767 19416 | 7035 | 0.034 | -0.0724 | No | ||
| 49 | GCNT2 | 9168 3166 3243 21661 3270 | 7156 | 0.032 | -0.0784 | No | ||
| 50 | PEX16 | 2681 | 7191 | 0.032 | -0.0798 | No | ||
| 51 | CHRDL1 | 8180 13504 | 7296 | 0.030 | -0.0850 | No | ||
| 52 | NKX2-8 | 9465 | 7383 | 0.028 | -0.0892 | No | ||
| 53 | LTBP1 | 1536 1604 6503 1559 1564 1518 | 7426 | 0.028 | -0.0910 | No | ||
| 54 | RHOA | 8624 4409 4410 | 7462 | 0.027 | -0.0925 | No | ||
| 55 | REPS1 | 3431 20087 | 7475 | 0.027 | -0.0928 | No | ||
| 56 | ANK2 | 4275 | 7516 | 0.026 | -0.0945 | No | ||
| 57 | KCNK7 | 23784 | 7772 | 0.023 | -0.1080 | No | ||
| 58 | AK2 | 8563 2479 16073 | 7871 | 0.022 | -0.1130 | No | ||
| 59 | HOXA3 | 9108 1075 | 7915 | 0.021 | -0.1150 | No | ||
| 60 | BHLHB5 | 15629 | 8036 | 0.019 | -0.1212 | No | ||
| 61 | DDR2 | 13767 | 8066 | 0.019 | -0.1225 | No | ||
| 62 | IGSF1 | 2589 2595 2588 2560 | 8159 | 0.018 | -0.1272 | No | ||
| 63 | CLDN8 | 22549 1747 | 8232 | 0.017 | -0.1308 | No | ||
| 64 | PFTK1 | 16924 | 8292 | 0.016 | -0.1338 | No | ||
| 65 | MB | 22232 | 8528 | 0.014 | -0.1463 | No | ||
| 66 | FBXL14 | 4155 | 8554 | 0.013 | -0.1474 | No | ||
| 67 | RHOV | 14470 | 8875 | 0.010 | -0.1646 | No | ||
| 68 | FSTL5 | 10018 | 8934 | 0.009 | -0.1676 | No | ||
| 69 | ASPA | 20354 | 8954 | 0.009 | -0.1685 | No | ||
| 70 | NEUROD6 | 17138 | 9111 | 0.007 | -0.1768 | No | ||
| 71 | SLC39A4 | 22240 2281 | 9435 | 0.004 | -0.1943 | No | ||
| 72 | UBE2S | 17979 | 9587 | 0.002 | -0.2024 | No | ||
| 73 | NR3C2 | 18562 13798 | 9628 | 0.002 | -0.2046 | No | ||
| 74 | DACT1 | 7203 | 9740 | 0.001 | -0.2106 | No | ||
| 75 | FGF12 | 1723 22621 | 9778 | 0.000 | -0.2126 | No | ||
| 76 | RAPSN | 2879 14951 | 9857 | -0.001 | -0.2168 | No | ||
| 77 | IVL | 15263 | 9964 | -0.002 | -0.2225 | No | ||
| 78 | B3GALT2 | 6498 | 10027 | -0.003 | -0.2258 | No | ||
| 79 | CAPZA1 | 8687 | 10088 | -0.003 | -0.2290 | No | ||
| 80 | FBXO24 | 12918 3649 | 10363 | -0.006 | -0.2438 | No | ||
| 81 | SPOCK2 | 13585 | 10449 | -0.007 | -0.2483 | No | ||
| 82 | HOXB8 | 9111 | 10477 | -0.008 | -0.2496 | No | ||
| 83 | TGM5 | 2687 14460 | 10643 | -0.009 | -0.2584 | No | ||
| 84 | RHOC | 15467 | 10677 | -0.009 | -0.2600 | No | ||
| 85 | GNG3 | 23752 | 10787 | -0.011 | -0.2658 | No | ||
| 86 | PIP5K2B | 20269 1254 | 10849 | -0.011 | -0.2689 | No | ||
| 87 | ALOX12B | 20825 | 10896 | -0.012 | -0.2712 | No | ||
| 88 | CDH13 | 4506 3826 | 11044 | -0.014 | -0.2790 | No | ||
| 89 | RAB3C | 21351 | 11691 | -0.022 | -0.3137 | No | ||
| 90 | GRIN2A | 22668 | 11864 | -0.024 | -0.3226 | No | ||
| 91 | ZYX | 10443 | 12079 | -0.026 | -0.3338 | No | ||
| 92 | CNTF | 3741 8761 3680 3736 | 12085 | -0.026 | -0.3337 | No | ||
| 93 | BDNF | 14926 2797 | 12118 | -0.027 | -0.3350 | No | ||
| 94 | HOXB3 | 20685 | 12141 | -0.027 | -0.3358 | No | ||
| 95 | ODF2 | 9501 5205 2862 2949 | 12246 | -0.029 | -0.3410 | No | ||
| 96 | FGF16 | 24270 | 12320 | -0.029 | -0.3445 | No | ||
| 97 | JPH3 | 18441 | 12510 | -0.032 | -0.3542 | No | ||
| 98 | FBXL2 | 7793 | 12665 | -0.035 | -0.3620 | No | ||
| 99 | STC2 | 5526 | 12855 | -0.038 | -0.3717 | No | ||
| 100 | RFX4 | 7721 3410 | 12936 | -0.040 | -0.3754 | No | ||
| 101 | ADAM2 | 21777 | 13010 | -0.041 | -0.3787 | No | ||
| 102 | NOTCH2 | 15485 | 13013 | -0.041 | -0.3782 | No | ||
| 103 | CHN1 | 4246 | 13110 | -0.043 | -0.3827 | No | ||
| 104 | HNRPF | 13607 | 13113 | -0.043 | -0.3822 | No | ||
| 105 | CITED2 | 5118 14477 | 13366 | -0.048 | -0.3951 | No | ||
| 106 | SESN3 | 19561 | 13384 | -0.048 | -0.3953 | No | ||
| 107 | HNRPUL1 | 6118 17923 1608 10577 1345 | 13621 | -0.054 | -0.4073 | No | ||
| 108 | CDCA3 | 17284 | 13629 | -0.054 | -0.4068 | No | ||
| 109 | ARHGAP12 | 13257 | 13639 | -0.054 | -0.4065 | No | ||
| 110 | FMOD | 14133 | 13740 | -0.057 | -0.4110 | No | ||
| 111 | PCSK1N | 6624 | 13869 | -0.060 | -0.4170 | No | ||
| 112 | PFN2 | 15337 1818 | 14042 | -0.065 | -0.4253 | No | ||
| 113 | RARG | 22113 | 14166 | -0.069 | -0.4309 | No | ||
| 114 | ESRRG | 6447 | 14257 | -0.072 | -0.4347 | No | ||
| 115 | RGN | 24371 | 14331 | -0.075 | -0.4375 | No | ||
| 116 | FRAS1 | 16788 3589 | 14439 | -0.079 | -0.4421 | No | ||
| 117 | LRFN5 | 6213 | 14610 | -0.086 | -0.4500 | No | ||
| 118 | C6 | 22522 8671 | 14679 | -0.090 | -0.4523 | No | ||
| 119 | SDC1 | 5560 9953 | 14765 | -0.094 | -0.4554 | No | ||
| 120 | MAP4K4 | 11241 | 14782 | -0.095 | -0.4548 | No | ||
| 121 | ANKRD17 | 13496 3474 8170 | 14960 | -0.107 | -0.4628 | No | ||
| 122 | CACNB2 | 4466 8678 | 15085 | -0.118 | -0.4677 | No | ||
| 123 | JARID2 | 9200 | 15250 | -0.131 | -0.4745 | No | ||
| 124 | MAMDC1 | 21054 6798 | 15323 | -0.139 | -0.4763 | No | ||
| 125 | HOXB5 | 20687 | 15386 | -0.145 | -0.4774 | No | ||
| 126 | GRIA3 | 24349 11996 2649 | 15389 | -0.146 | -0.4753 | No | ||
| 127 | CD44 | 2881 2847 2893 4499 2724 | 15399 | -0.147 | -0.4735 | No | ||
| 128 | TENC1 | 22349 9927 | 15586 | -0.171 | -0.4810 | No | ||
| 129 | PRICKLE1 | 8379 | 15622 | -0.176 | -0.4801 | No | ||
| 130 | MGLL | 17368 1145 | 15678 | -0.185 | -0.4803 | No | ||
| 131 | SEMA7A | 19426 9803 | 15940 | -0.238 | -0.4908 | Yes | ||
| 132 | PCDH9 | 21736 | 16008 | -0.253 | -0.4905 | Yes | ||
| 133 | RIN1 | 10349 | 16024 | -0.258 | -0.4873 | Yes | ||
| 134 | ING3 | 17514 1019 | 16093 | -0.275 | -0.4868 | Yes | ||
| 135 | STK11IP | 14212 | 16162 | -0.295 | -0.4859 | Yes | ||
| 136 | EDG5 | 19216 | 16248 | -0.318 | -0.4857 | Yes | ||
| 137 | TYSND1 | 20007 | 16345 | -0.348 | -0.4855 | Yes | ||
| 138 | SNRPD1 | 23622 | 16359 | -0.353 | -0.4808 | Yes | ||
| 139 | UGCGL1 | 6721 | 16462 | -0.394 | -0.4802 | Yes | ||
| 140 | BSCL2 | 23940 | 16657 | -0.466 | -0.4836 | Yes | ||
| 141 | NR6A1 | 14596 | 16809 | -0.540 | -0.4834 | Yes | ||
| 142 | GTF3C2 | 7749 | 17120 | -0.721 | -0.4892 | Yes | ||
| 143 | SLC16A6 | 4186 | 17201 | -0.783 | -0.4814 | Yes | ||
| 144 | CDK2 | 3438 3373 19592 3322 | 17283 | -0.848 | -0.4728 | Yes | ||
| 145 | ARHGEF6 | 13297 13104 | 17385 | -0.926 | -0.4640 | Yes | ||
| 146 | YPEL1 | 22840 | 17387 | -0.929 | -0.4498 | Yes | ||
| 147 | PAFAH1B1 | 1340 5220 9524 | 17497 | -1.002 | -0.4403 | Yes | ||
| 148 | HOXB4 | 20686 | 17584 | -1.062 | -0.4286 | Yes | ||
| 149 | SSBP2 | 3171 7364 3252 | 17932 | -1.419 | -0.4256 | Yes | ||
| 150 | PHC1 | 17013 | 18003 | -1.513 | -0.4061 | Yes | ||
| 151 | PTPN22 | 1775 15470 | 18055 | -1.578 | -0.3845 | Yes | ||
| 152 | TCTA | 4167 | 18063 | -1.581 | -0.3606 | Yes | ||
| 153 | IGFBP4 | 20670 9160 | 18114 | -1.639 | -0.3381 | Yes | ||
| 154 | PREB | 6948 3628 | 18119 | -1.645 | -0.3130 | Yes | ||
| 155 | MYO18A | 20766 | 18143 | -1.683 | -0.2884 | Yes | ||
| 156 | DLX1 | 14988 | 18255 | -1.899 | -0.2651 | Yes | ||
| 157 | HES6 | 13882 4058 | 18269 | -1.932 | -0.2361 | Yes | ||
| 158 | MBD3 | 19695 | 18326 | -2.073 | -0.2073 | Yes | ||
| 159 | CSAD | 10899 | 18544 | -3.090 | -0.1715 | Yes | ||
| 160 | UTX | 10266 2574 | 18546 | -3.097 | -0.1239 | Yes | ||
| 161 | SOCS2 | 5694 | 18614 | -8.294 | 0.0001 | Yes |