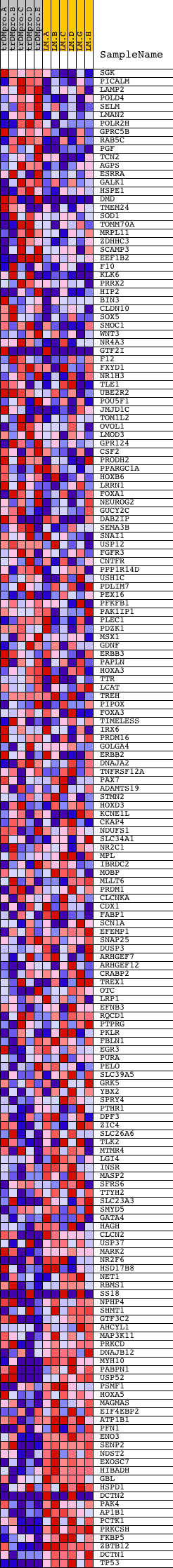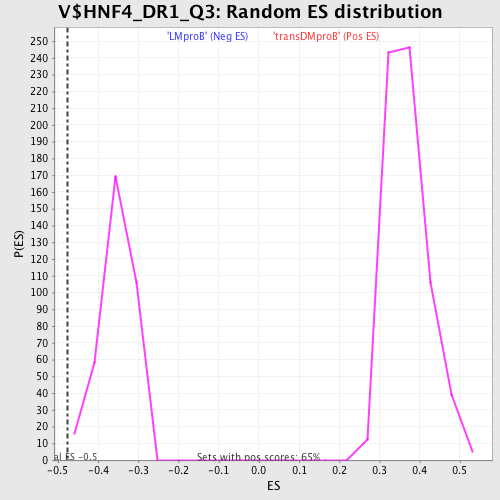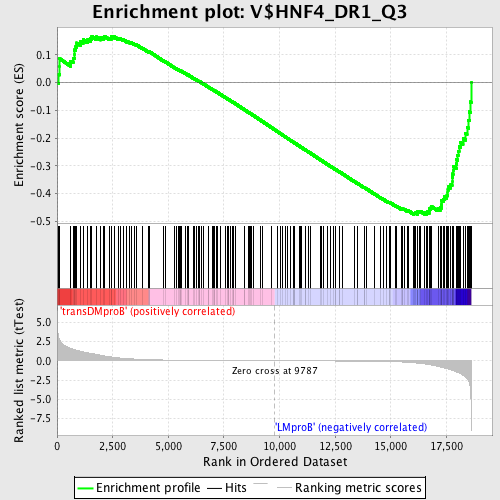
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | V$HNF4_DR1_Q3 |
| Enrichment Score (ES) | -0.47537664 |
| Normalized Enrichment Score (NES) | -1.3444158 |
| Nominal p-value | 0.0057306592 |
| FDR q-value | 0.51755357 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SGK | 9809 3419 | 69 | 2.979 | 0.0296 | No | ||
| 2 | PICALM | 18191 | 113 | 2.628 | 0.0567 | No | ||
| 3 | LAMP2 | 9267 2653 | 118 | 2.612 | 0.0857 | No | ||
| 4 | POLD4 | 12822 | 609 | 1.619 | 0.0773 | No | ||
| 5 | SELM | 20967 | 720 | 1.509 | 0.0882 | No | ||
| 6 | LMAN2 | 21454 | 774 | 1.455 | 0.1016 | No | ||
| 7 | POLR2H | 10888 | 779 | 1.450 | 0.1177 | No | ||
| 8 | GPRC5B | 17662 | 843 | 1.395 | 0.1299 | No | ||
| 9 | RAB5C | 20226 | 887 | 1.356 | 0.1427 | No | ||
| 10 | PGF | 21020 | 1054 | 1.230 | 0.1475 | No | ||
| 11 | TCN2 | 20550 | 1166 | 1.158 | 0.1544 | No | ||
| 12 | AGPS | 14979 | 1381 | 1.008 | 0.1541 | No | ||
| 13 | ESRRA | 23802 | 1500 | 0.971 | 0.1586 | No | ||
| 14 | GALK1 | 20147 | 1561 | 0.939 | 0.1659 | No | ||
| 15 | HSPE1 | 9133 | 1756 | 0.828 | 0.1646 | No | ||
| 16 | DMD | 24295 2647 | 1963 | 0.710 | 0.1614 | No | ||
| 17 | TMEM24 | 19150 | 2079 | 0.652 | 0.1625 | No | ||
| 18 | SOD1 | 9846 | 2119 | 0.635 | 0.1675 | No | ||
| 19 | TOMM70A | 6608 | 2368 | 0.515 | 0.1598 | No | ||
| 20 | MRPL11 | 23774 | 2428 | 0.495 | 0.1621 | No | ||
| 21 | ZDHHC3 | 7598 12763 | 2434 | 0.493 | 0.1674 | No | ||
| 22 | SCAMP3 | 15543 | 2565 | 0.442 | 0.1653 | No | ||
| 23 | EEF1B2 | 4131 12063 | 2779 | 0.369 | 0.1578 | No | ||
| 24 | F10 | 18682 | 2838 | 0.353 | 0.1587 | No | ||
| 25 | KLK6 | 9231 9626 | 2971 | 0.316 | 0.1550 | No | ||
| 26 | PRRX2 | 15047 | 3140 | 0.278 | 0.1490 | No | ||
| 27 | HIP2 | 3626 3485 16838 | 3261 | 0.254 | 0.1454 | No | ||
| 28 | BIN3 | 21969 | 3329 | 0.242 | 0.1445 | No | ||
| 29 | CLDN10 | 21935 2785 2601 2720 7188 | 3492 | 0.215 | 0.1381 | No | ||
| 30 | SOX5 | 9850 1044 5480 16937 | 3554 | 0.205 | 0.1371 | No | ||
| 31 | SMOC1 | 21221 | 3835 | 0.171 | 0.1238 | No | ||
| 32 | WNT3 | 20635 | 4109 | 0.141 | 0.1106 | No | ||
| 33 | NR4A3 | 9473 16212 5183 | 4142 | 0.138 | 0.1104 | No | ||
| 34 | GTF2I | 16353 5491 9863 3280 3184 | 4155 | 0.137 | 0.1113 | No | ||
| 35 | F12 | 3260 21453 | 4780 | 0.095 | 0.0785 | No | ||
| 36 | FXYD1 | 17874 85 | 4860 | 0.092 | 0.0752 | No | ||
| 37 | NR1H3 | 10258 2787 | 5262 | 0.075 | 0.0543 | No | ||
| 38 | TLE1 | 5765 10185 15859 | 5375 | 0.072 | 0.0491 | No | ||
| 39 | UBE2R2 | 16240 | 5466 | 0.069 | 0.0450 | No | ||
| 40 | POU5F1 | 1547 23255 | 5511 | 0.067 | 0.0433 | No | ||
| 41 | JMJD1C | 11531 19996 | 5556 | 0.066 | 0.0417 | No | ||
| 42 | TOM1L2 | 10115 | 5559 | 0.066 | 0.0423 | No | ||
| 43 | OVOL1 | 23983 | 5603 | 0.065 | 0.0407 | No | ||
| 44 | LMOD3 | 17059 | 5757 | 0.061 | 0.0331 | No | ||
| 45 | GPR124 | 18643 918 | 5862 | 0.058 | 0.0281 | No | ||
| 46 | CSF2 | 20454 | 5887 | 0.057 | 0.0274 | No | ||
| 47 | PRODH2 | 1854 18300 | 5888 | 0.057 | 0.0281 | No | ||
| 48 | PPARGC1A | 16533 | 6137 | 0.051 | 0.0152 | No | ||
| 49 | HOXB6 | 20688 | 6165 | 0.050 | 0.0143 | No | ||
| 50 | LRRN1 | 17343 | 6276 | 0.048 | 0.0088 | No | ||
| 51 | FOXA1 | 21060 | 6363 | 0.046 | 0.0047 | No | ||
| 52 | NEUROG2 | 15426 | 6383 | 0.045 | 0.0042 | No | ||
| 53 | GUCY2C | 1140 16954 | 6417 | 0.045 | 0.0029 | No | ||
| 54 | DAB2IP | 12808 2692 | 6495 | 0.043 | -0.0008 | No | ||
| 55 | SEMA3B | 5422 | 6572 | 0.042 | -0.0045 | No | ||
| 56 | SNAI1 | 14726 | 6817 | 0.037 | -0.0173 | No | ||
| 57 | USP12 | 5825 5824 | 6818 | 0.037 | -0.0169 | No | ||
| 58 | FGFR3 | 8969 3566 | 6996 | 0.035 | -0.0261 | No | ||
| 59 | CNTFR | 2515 15906 | 7038 | 0.034 | -0.0279 | No | ||
| 60 | PPP1R14D | 14471 | 7086 | 0.033 | -0.0301 | No | ||
| 61 | USH1C | 1471 17821 | 7088 | 0.033 | -0.0298 | No | ||
| 62 | PDLIM7 | 7434 | 7180 | 0.032 | -0.0344 | No | ||
| 63 | PEX16 | 2681 | 7191 | 0.032 | -0.0345 | No | ||
| 64 | PFKFB1 | 9552 | 7345 | 0.029 | -0.0425 | No | ||
| 65 | PAK1IP1 | 21660 | 7567 | 0.026 | -0.0542 | No | ||
| 66 | PLEC1 | 2273 2198 2205 2178 2230 22247 2213 2231 2295 2172 5263 2266 | 7637 | 0.025 | -0.0577 | No | ||
| 67 | PDZK1 | 15490 | 7692 | 0.024 | -0.0603 | No | ||
| 68 | MSX1 | 16547 | 7714 | 0.024 | -0.0612 | No | ||
| 69 | GDNF | 22514 2275 | 7809 | 0.022 | -0.0661 | No | ||
| 70 | ERBB3 | 19593 | 7882 | 0.021 | -0.0697 | No | ||
| 71 | PAPLN | 9301 | 7913 | 0.021 | -0.0711 | No | ||
| 72 | HOXA3 | 9108 1075 | 7915 | 0.021 | -0.0709 | No | ||
| 73 | TTR | 23615 | 8022 | 0.019 | -0.0765 | No | ||
| 74 | LCAT | 18762 | 8410 | 0.015 | -0.0973 | No | ||
| 75 | TREH | 19474 | 8414 | 0.015 | -0.0973 | No | ||
| 76 | PIPOX | 20340 | 8616 | 0.013 | -0.1081 | No | ||
| 77 | FOXA3 | 17949 | 8662 | 0.012 | -0.1104 | No | ||
| 78 | TIMELESS | 10177 | 8686 | 0.012 | -0.1115 | No | ||
| 79 | IRX6 | 7225 | 8737 | 0.011 | -0.1141 | No | ||
| 80 | PRDM16 | 12893 | 8809 | 0.011 | -0.1178 | No | ||
| 81 | GOLGA4 | 12022 | 9132 | 0.007 | -0.1352 | No | ||
| 82 | ERBB2 | 8913 | 9243 | 0.006 | -0.1411 | No | ||
| 83 | DNAJA2 | 12133 18806 | 9629 | 0.002 | -0.1619 | No | ||
| 84 | TNFRSF12A | 23107 | 9919 | -0.002 | -0.1776 | No | ||
| 85 | PAX7 | 15697 | 10053 | -0.003 | -0.1848 | No | ||
| 86 | ADAMTS19 | 23544 | 10134 | -0.004 | -0.1891 | No | ||
| 87 | STMN2 | 15638 | 10263 | -0.005 | -0.1959 | No | ||
| 88 | HOXD3 | 9115 | 10347 | -0.006 | -0.2004 | No | ||
| 89 | KCNE1L | 24044 | 10506 | -0.008 | -0.2089 | No | ||
| 90 | CKAP4 | 19663 | 10631 | -0.009 | -0.2155 | No | ||
| 91 | NDUFS1 | 13940 | 10640 | -0.009 | -0.2158 | No | ||
| 92 | SLC34A1 | 9824 | 10655 | -0.009 | -0.2165 | No | ||
| 93 | NR2C1 | 10215 | 10670 | -0.009 | -0.2171 | No | ||
| 94 | MPL | 15780 | 10903 | -0.012 | -0.2296 | No | ||
| 95 | IBRDC2 | 5747 | 10917 | -0.012 | -0.2301 | No | ||
| 96 | MOBP | 5112 | 10988 | -0.013 | -0.2338 | No | ||
| 97 | MLLT6 | 20678 | 11145 | -0.015 | -0.2421 | No | ||
| 98 | PRDM1 | 19775 3337 | 11162 | -0.015 | -0.2428 | No | ||
| 99 | CLCNKA | 15690 | 11301 | -0.016 | -0.2501 | No | ||
| 100 | CDX1 | 23429 | 11372 | -0.017 | -0.2537 | No | ||
| 101 | FABP1 | 17420 | 11830 | -0.023 | -0.2782 | No | ||
| 102 | SCN1A | 9784 | 11868 | -0.024 | -0.2800 | No | ||
| 103 | EFEMP1 | 20937 | 11978 | -0.025 | -0.2856 | No | ||
| 104 | SNAP25 | 5466 | 12150 | -0.027 | -0.2946 | No | ||
| 105 | DUSP3 | 1358 13022 | 12297 | -0.029 | -0.3022 | No | ||
| 106 | ARHGEF7 | 3823 | 12421 | -0.031 | -0.3085 | No | ||
| 107 | ARHGEF12 | 19156 | 12493 | -0.032 | -0.3120 | No | ||
| 108 | CRABP2 | 15554 | 12504 | -0.032 | -0.3122 | No | ||
| 109 | TREX1 | 10219 3111 | 12675 | -0.035 | -0.3210 | No | ||
| 110 | OTC | 9518 | 12703 | -0.035 | -0.3220 | No | ||
| 111 | LRP1 | 9284 | 12843 | -0.038 | -0.3292 | No | ||
| 112 | EFNB3 | 20391 | 13374 | -0.048 | -0.3574 | No | ||
| 113 | RQCD1 | 12205 | 13516 | -0.051 | -0.3644 | No | ||
| 114 | PTPRG | 22097 3151 | 13521 | -0.051 | -0.3641 | No | ||
| 115 | PKLR | 1850 15545 | 13808 | -0.059 | -0.3789 | No | ||
| 116 | FBLN1 | 4716 2208 | 13826 | -0.059 | -0.3792 | No | ||
| 117 | EGR3 | 4656 | 13898 | -0.061 | -0.3824 | No | ||
| 118 | PURA | 9670 | 14274 | -0.073 | -0.4019 | No | ||
| 119 | PELO | 8360 | 14521 | -0.082 | -0.4143 | No | ||
| 120 | SLC39A5 | 3364 19596 | 14663 | -0.089 | -0.4210 | No | ||
| 121 | GRK5 | 9036 23814 | 14825 | -0.097 | -0.4286 | No | ||
| 122 | YBX2 | 20818 | 14922 | -0.105 | -0.4326 | No | ||
| 123 | SPRY4 | 23448 | 14931 | -0.106 | -0.4319 | No | ||
| 124 | PTHR1 | 5318 18985 | 15004 | -0.111 | -0.4345 | No | ||
| 125 | DPF3 | 7659 12853 | 15192 | -0.126 | -0.4433 | No | ||
| 126 | ZIC4 | 5992 | 15258 | -0.132 | -0.4453 | No | ||
| 127 | SLC26A6 | 19304 | 15477 | -0.156 | -0.4554 | No | ||
| 128 | TLK2 | 20628 | 15521 | -0.160 | -0.4559 | No | ||
| 129 | MTMR4 | 20712 | 15544 | -0.164 | -0.4553 | No | ||
| 130 | LGI4 | 18296 | 15600 | -0.173 | -0.4563 | No | ||
| 131 | INSR | 18950 | 15769 | -0.203 | -0.4632 | No | ||
| 132 | MASP2 | 2547 | 15791 | -0.207 | -0.4620 | No | ||
| 133 | SFRS6 | 14751 | 16039 | -0.262 | -0.4724 | Yes | ||
| 134 | TTYH2 | 8585 4382 20605 | 16058 | -0.266 | -0.4704 | Yes | ||
| 135 | SLC23A3 | 10367 | 16099 | -0.277 | -0.4695 | Yes | ||
| 136 | SMYD5 | 17386 | 16189 | -0.303 | -0.4709 | Yes | ||
| 137 | GATA4 | 21792 4755 | 16200 | -0.306 | -0.4681 | Yes | ||
| 138 | HAGH | 9020 | 16209 | -0.307 | -0.4651 | Yes | ||
| 139 | CLCN2 | 22637 | 16288 | -0.330 | -0.4656 | Yes | ||
| 140 | USP37 | 13923 | 16324 | -0.341 | -0.4637 | Yes | ||
| 141 | MARK2 | 8899 | 16521 | -0.412 | -0.4697 | Yes | ||
| 142 | NR2F6 | 4651 8876 18848 8912 | 16615 | -0.444 | -0.4698 | Yes | ||
| 143 | HSD17B8 | 9061 | 16635 | -0.451 | -0.4657 | Yes | ||
| 144 | NET1 | 7100 | 16719 | -0.498 | -0.4647 | Yes | ||
| 145 | RBMS1 | 14580 | 16721 | -0.499 | -0.4591 | Yes | ||
| 146 | SS18 | 6506 6505 2030 | 16747 | -0.513 | -0.4547 | Yes | ||
| 147 | NPHP4 | 15978 | 16804 | -0.537 | -0.4518 | Yes | ||
| 148 | SHMT1 | 5431 | 16818 | -0.543 | -0.4464 | Yes | ||
| 149 | GTF3C2 | 7749 | 17120 | -0.721 | -0.4546 | Yes | ||
| 150 | AHCYL1 | 6037 | 17216 | -0.800 | -0.4508 | Yes | ||
| 151 | MAP3K11 | 11163 | 17263 | -0.835 | -0.4440 | Yes | ||
| 152 | PRKCD | 21897 | 17286 | -0.851 | -0.4356 | Yes | ||
| 153 | DNAJB12 | 7146 | 17289 | -0.854 | -0.4262 | Yes | ||
| 154 | MYH10 | 8077 | 17365 | -0.915 | -0.4200 | Yes | ||
| 155 | PABPN1 | 12018 | 17408 | -0.946 | -0.4117 | Yes | ||
| 156 | USP52 | 3397 19841 | 17522 | -1.014 | -0.4065 | Yes | ||
| 157 | PSMF1 | 6009 | 17566 | -1.048 | -0.3971 | Yes | ||
| 158 | HOXA5 | 17149 | 17569 | -1.050 | -0.3854 | Yes | ||
| 159 | MAGMAS | 1677 22678 | 17606 | -1.083 | -0.3752 | Yes | ||
| 160 | EIF4EBP2 | 4662 | 17698 | -1.180 | -0.3670 | Yes | ||
| 161 | ATP1B1 | 4420 | 17752 | -1.235 | -0.3560 | Yes | ||
| 162 | PFN1 | 9555 | 17756 | -1.238 | -0.3423 | Yes | ||
| 163 | ENO3 | 8905 | 17777 | -1.259 | -0.3293 | Yes | ||
| 164 | SENP2 | 7990 | 17798 | -1.280 | -0.3160 | Yes | ||
| 165 | NDST2 | 21906 | 17806 | -1.288 | -0.3020 | Yes | ||
| 166 | EXOSC7 | 19256 | 17939 | -1.431 | -0.2931 | Yes | ||
| 167 | HIBADH | 17144 | 17956 | -1.452 | -0.2777 | Yes | ||
| 168 | GBL | 23091 1612 | 18005 | -1.522 | -0.2633 | Yes | ||
| 169 | HSPD1 | 4078 9129 | 18026 | -1.548 | -0.2471 | Yes | ||
| 170 | DCTN2 | 7635 12812 | 18067 | -1.582 | -0.2315 | Yes | ||
| 171 | PAK4 | 17909 | 18131 | -1.667 | -0.2163 | Yes | ||
| 172 | AP1B1 | 8599 | 18245 | -1.884 | -0.2013 | Yes | ||
| 173 | PCTK1 | 24369 | 18357 | -2.140 | -0.1834 | Yes | ||
| 174 | PRKCSH | 19535 | 18426 | -2.304 | -0.1612 | Yes | ||
| 175 | FKBP5 | 23051 | 18492 | -2.579 | -0.1359 | Yes | ||
| 176 | ZBTB12 | 23268 | 18529 | -2.888 | -0.1055 | Yes | ||
| 177 | DCTN1 | 1100 17392 | 18572 | -3.406 | -0.0696 | Yes | ||
| 178 | TP53 | 20822 | 18610 | -6.426 | 0.0003 | Yes |