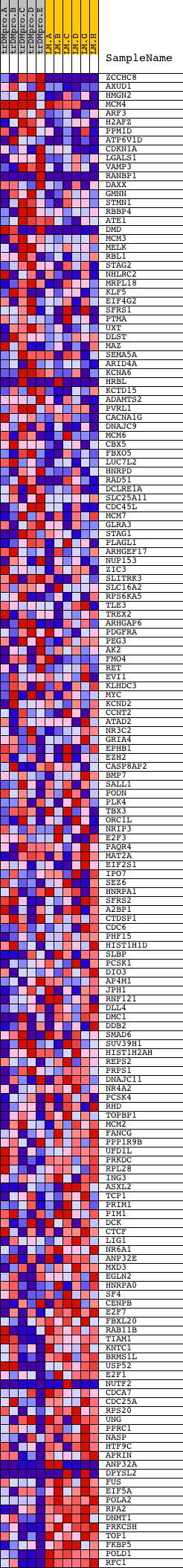
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
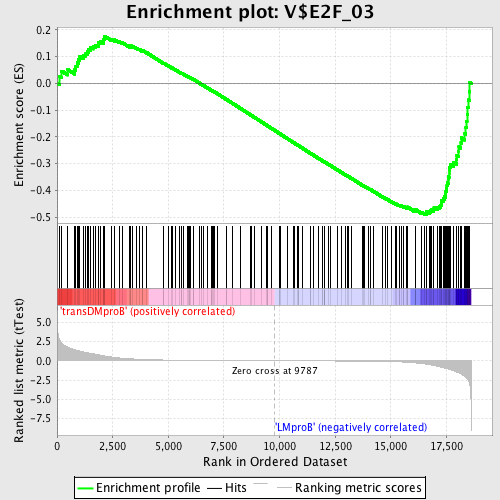
| Upregulated in class | LMproB |
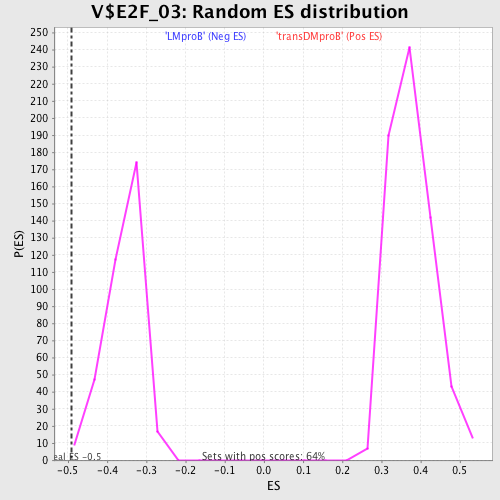
| GeneSet | V$E2F_03 |
| Enrichment Score (ES) | -0.48988375 |
| Normalized Enrichment Score (NES) | -1.3735356 |
| Nominal p-value | 0.0054945056 |
| FDR q-value | 0.6097444 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ZCCHC8 | 3570 7696 | 109 | 2.662 | 0.0249 | No | ||
| 2 | AXUD1 | 18967 | 204 | 2.257 | 0.0459 | No | ||
| 3 | HMGN2 | 9095 | 481 | 1.743 | 0.0511 | No | ||
| 4 | MCM4 | 22655 1708 | 798 | 1.434 | 0.0505 | No | ||
| 5 | ARF3 | 22137 | 842 | 1.396 | 0.0643 | No | ||
| 6 | H2AFZ | 6957 | 901 | 1.341 | 0.0767 | No | ||
| 7 | PPM1D | 20721 | 956 | 1.299 | 0.0888 | No | ||
| 8 | ATP6V1D | 7872 | 1021 | 1.251 | 0.0998 | No | ||
| 9 | CDKN1A | 4511 8729 | 1187 | 1.141 | 0.1040 | No | ||
| 10 | LGALS1 | 22429 | 1293 | 1.071 | 0.1107 | No | ||
| 11 | VAMP3 | 15660 | 1351 | 1.026 | 0.1195 | No | ||
| 12 | RANBP1 | 9692 5357 | 1416 | 1.000 | 0.1276 | No | ||
| 13 | DAXX | 23290 | 1495 | 0.973 | 0.1346 | No | ||
| 14 | GMNN | 21513 | 1618 | 0.906 | 0.1385 | No | ||
| 15 | STMN1 | 9261 | 1734 | 0.837 | 0.1420 | No | ||
| 16 | RBBP4 | 5364 | 1852 | 0.772 | 0.1445 | No | ||
| 17 | ATE1 | 388 17602 | 1865 | 0.764 | 0.1527 | No | ||
| 18 | DMD | 24295 2647 | 1963 | 0.710 | 0.1557 | No | ||
| 19 | MCM3 | 13991 | 2074 | 0.654 | 0.1573 | No | ||
| 20 | MELK | 2453 16227 | 2082 | 0.652 | 0.1644 | No | ||
| 21 | RBL1 | 14373 | 2123 | 0.632 | 0.1696 | No | ||
| 22 | STAG2 | 5521 | 2136 | 0.623 | 0.1761 | No | ||
| 23 | NHLRC2 | 23828 | 2445 | 0.488 | 0.1651 | No | ||
| 24 | MRPL18 | 12567 | 2593 | 0.431 | 0.1621 | No | ||
| 25 | KLF5 | 4456 | 2802 | 0.361 | 0.1550 | No | ||
| 26 | EIF4G2 | 1908 8892 | 2927 | 0.331 | 0.1521 | No | ||
| 27 | SFRS1 | 8492 | 3253 | 0.256 | 0.1374 | No | ||
| 28 | PTMA | 9657 | 3303 | 0.245 | 0.1376 | No | ||
| 29 | UXT | 5837 10267 | 3309 | 0.245 | 0.1402 | No | ||
| 30 | DLST | 8131 | 3410 | 0.228 | 0.1374 | No | ||
| 31 | MAZ | 1327 17623 | 3581 | 0.201 | 0.1305 | No | ||
| 32 | SEMA5A | 22496 5423 | 3694 | 0.188 | 0.1266 | No | ||
| 33 | ARID4A | 2080 6215 | 3819 | 0.173 | 0.1219 | No | ||
| 34 | KCNA6 | 664 16990 665 4938 | 3826 | 0.172 | 0.1236 | No | ||
| 35 | HRBL | 3452 16326 3614 | 4025 | 0.149 | 0.1146 | No | ||
| 36 | KCTD15 | 17864 | 4771 | 0.096 | 0.0753 | No | ||
| 37 | ADAMTS2 | 5706 20899 1356 | 4791 | 0.095 | 0.0754 | No | ||
| 38 | PVRL1 | 3006 19482 12211 7192 | 4991 | 0.086 | 0.0656 | No | ||
| 39 | CACNA1G | 1237 940 20292 | 5162 | 0.079 | 0.0573 | No | ||
| 40 | DNAJC9 | 21912 | 5182 | 0.079 | 0.0571 | No | ||
| 41 | MCM6 | 4000 13845 4119 | 5338 | 0.073 | 0.0496 | No | ||
| 42 | CBX5 | 22108 4485 | 5512 | 0.067 | 0.0410 | No | ||
| 43 | FBXO5 | 19830 | 5577 | 0.066 | 0.0383 | No | ||
| 44 | LUC7L2 | 5313 5312 | 5596 | 0.065 | 0.0380 | No | ||
| 45 | HNRPD | 8645 | 5697 | 0.062 | 0.0333 | No | ||
| 46 | RAD51 | 2897 14903 | 5880 | 0.057 | 0.0241 | No | ||
| 47 | DCLRE1A | 23641 | 5896 | 0.057 | 0.0240 | No | ||
| 48 | SLC25A11 | 20366 1368 | 5952 | 0.056 | 0.0216 | No | ||
| 49 | CDC45L | 22642 1752 | 5968 | 0.055 | 0.0215 | No | ||
| 50 | MCM7 | 9372 3568 | 5999 | 0.054 | 0.0205 | No | ||
| 51 | GLRA3 | 18617 | 6108 | 0.052 | 0.0152 | No | ||
| 52 | STAG1 | 5520 9905 2987 | 6133 | 0.051 | 0.0145 | No | ||
| 53 | PLAGL1 | 10372 3414 | 6386 | 0.045 | 0.0014 | No | ||
| 54 | ARHGEF17 | 17737 | 6496 | 0.043 | -0.0040 | No | ||
| 55 | NUP153 | 21474 | 6570 | 0.042 | -0.0075 | No | ||
| 56 | ZIC3 | 10430 | 6781 | 0.038 | -0.0185 | No | ||
| 57 | SLITRK3 | 11816 | 6922 | 0.036 | -0.0256 | No | ||
| 58 | SLC16A2 | 24086 | 6943 | 0.035 | -0.0263 | No | ||
| 59 | RPS6KA5 | 2076 21005 | 6994 | 0.035 | -0.0286 | No | ||
| 60 | TLE3 | 5767 19416 | 7035 | 0.034 | -0.0304 | No | ||
| 61 | TREX2 | 24137 | 7071 | 0.033 | -0.0319 | No | ||
| 62 | ARHGAP6 | 24207 | 7219 | 0.031 | -0.0395 | No | ||
| 63 | PDGFRA | 16824 | 7615 | 0.025 | -0.0606 | No | ||
| 64 | PEG3 | 5240 | 7631 | 0.025 | -0.0612 | No | ||
| 65 | AK2 | 8563 2479 16073 | 7871 | 0.022 | -0.0739 | No | ||
| 66 | FMO4 | 13786 | 8233 | 0.017 | -0.0932 | No | ||
| 67 | RET | 17028 | 8711 | 0.012 | -0.1190 | No | ||
| 68 | EVI1 | 4689 8926 | 8754 | 0.011 | -0.1211 | No | ||
| 69 | KLHDC3 | 22956 | 8860 | 0.010 | -0.1267 | No | ||
| 70 | MYC | 22465 9435 | 9165 | 0.007 | -0.1431 | No | ||
| 71 | KCND2 | 4941 17515 | 9403 | 0.004 | -0.1559 | No | ||
| 72 | CCNT2 | 3993 13079 | 9444 | 0.004 | -0.1580 | No | ||
| 73 | ATAD2 | 2268 2256 | 9471 | 0.003 | -0.1594 | No | ||
| 74 | NR3C2 | 18562 13798 | 9628 | 0.002 | -0.1678 | No | ||
| 75 | GRIA4 | 19249 | 9989 | -0.002 | -0.1873 | No | ||
| 76 | EPHB1 | 19024 | 9997 | -0.003 | -0.1876 | No | ||
| 77 | EZH2 | 1092 17163 | 10050 | -0.003 | -0.1904 | No | ||
| 78 | CASP8AP2 | 16253 | 10349 | -0.006 | -0.2065 | No | ||
| 79 | BMP7 | 14324 | 10622 | -0.009 | -0.2211 | No | ||
| 80 | SALL1 | 18796 12207 | 10682 | -0.009 | -0.2242 | No | ||
| 81 | PODN | 15811 | 10791 | -0.011 | -0.2300 | No | ||
| 82 | PLK4 | 1870 1874 | 10857 | -0.011 | -0.2333 | No | ||
| 83 | TBX3 | 16723 | 10868 | -0.012 | -0.2338 | No | ||
| 84 | ORC1L | 327 16144 | 11011 | -0.013 | -0.2413 | No | ||
| 85 | NRIP3 | 17679 | 11374 | -0.017 | -0.2607 | No | ||
| 86 | E2F3 | 21500 8874 | 11388 | -0.017 | -0.2612 | No | ||
| 87 | PAQR4 | 23105 | 11508 | -0.019 | -0.2675 | No | ||
| 88 | MAT2A | 10553 10554 6099 | 11769 | -0.022 | -0.2813 | No | ||
| 89 | EIF2S1 | 4658 | 11931 | -0.024 | -0.2897 | No | ||
| 90 | IPO7 | 6130 | 11947 | -0.025 | -0.2903 | No | ||
| 91 | SEZ6 | 20764 | 11999 | -0.025 | -0.2927 | No | ||
| 92 | HNRPA1 | 9103 4860 | 12007 | -0.025 | -0.2928 | No | ||
| 93 | SFRS2 | 9807 20136 | 12015 | -0.025 | -0.2929 | No | ||
| 94 | A2BP1 | 11212 1652 1689 | 12196 | -0.028 | -0.3023 | No | ||
| 95 | CTDSP1 | 4121 10407 | 12284 | -0.029 | -0.3067 | No | ||
| 96 | CDC6 | 6221 6220 | 12613 | -0.034 | -0.3241 | No | ||
| 97 | PHF15 | 20467 8050 | 12804 | -0.037 | -0.3340 | No | ||
| 98 | HIST1H1D | 4821 | 12974 | -0.040 | -0.3427 | No | ||
| 99 | SLBP | 9822 | 13033 | -0.042 | -0.3453 | No | ||
| 100 | PCSK1 | 21600 | 13091 | -0.043 | -0.3479 | No | ||
| 101 | DIO3 | 19359 | 13221 | -0.045 | -0.3544 | No | ||
| 102 | AP4M1 | 3499 16656 | 13709 | -0.056 | -0.3802 | No | ||
| 103 | JPH1 | 13994 | 13771 | -0.058 | -0.3828 | No | ||
| 104 | RNF121 | 7952 373 | 13813 | -0.059 | -0.3843 | No | ||
| 105 | DLL4 | 7040 | 14005 | -0.064 | -0.3939 | No | ||
| 106 | DMC1 | 22207 | 14082 | -0.066 | -0.3973 | No | ||
| 107 | DDB2 | 2721 2878 14525 2661 | 14086 | -0.067 | -0.3967 | No | ||
| 108 | SMAD6 | 19083 | 14205 | -0.071 | -0.4023 | No | ||
| 109 | SUV39H1 | 24194 | 14634 | -0.088 | -0.4245 | No | ||
| 110 | HIST1H2AH | 11414 | 14776 | -0.095 | -0.4310 | No | ||
| 111 | REPS2 | 24018 | 14838 | -0.099 | -0.4332 | No | ||
| 112 | PRPS1 | 24233 | 15031 | -0.113 | -0.4423 | No | ||
| 113 | DNAJC11 | 2486 6068 | 15215 | -0.128 | -0.4507 | No | ||
| 114 | NR4A2 | 5203 | 15242 | -0.131 | -0.4506 | No | ||
| 115 | PCSK4 | 19696 3296 3394 18705 | 15375 | -0.144 | -0.4561 | No | ||
| 116 | RHD | 16042 | 15384 | -0.145 | -0.4548 | No | ||
| 117 | TOPBP1 | 10659 | 15461 | -0.154 | -0.4572 | No | ||
| 118 | MCM2 | 17074 | 15571 | -0.167 | -0.4611 | No | ||
| 119 | FANCG | 15904 | 15580 | -0.169 | -0.4596 | No | ||
| 120 | PPP1R9B | 20698 | 15722 | -0.194 | -0.4650 | No | ||
| 121 | UFD1L | 22825 | 15737 | -0.198 | -0.4635 | No | ||
| 122 | PRKDC | 9617 22847 | 15750 | -0.199 | -0.4618 | No | ||
| 123 | RPL28 | 5392 | 16088 | -0.274 | -0.4769 | No | ||
| 124 | ING3 | 17514 1019 | 16093 | -0.275 | -0.4740 | No | ||
| 125 | ASXL2 | 7957 | 16110 | -0.280 | -0.4716 | No | ||
| 126 | TCP1 | 23380 1509 | 16384 | -0.365 | -0.4822 | No | ||
| 127 | PRIM1 | 19847 | 16527 | -0.414 | -0.4851 | Yes | ||
| 128 | PIM1 | 23308 | 16594 | -0.436 | -0.4836 | Yes | ||
| 129 | DCK | 16808 | 16607 | -0.441 | -0.4792 | Yes | ||
| 130 | CTCF | 18490 | 16750 | -0.514 | -0.4809 | Yes | ||
| 131 | LIG1 | 18388 1749 1493 | 16778 | -0.526 | -0.4763 | Yes | ||
| 132 | NR6A1 | 14596 | 16809 | -0.540 | -0.4717 | Yes | ||
| 133 | ANP32E | 15500 | 16925 | -0.598 | -0.4710 | Yes | ||
| 134 | MXD3 | 21455 | 16939 | -0.603 | -0.4648 | Yes | ||
| 135 | EGLN2 | 17920 | 17088 | -0.704 | -0.4646 | Yes | ||
| 136 | HNRPA0 | 13382 | 17172 | -0.759 | -0.4604 | Yes | ||
| 137 | SF4 | 18593 | 17247 | -0.823 | -0.4549 | Yes | ||
| 138 | CENPB | 14424 | 17282 | -0.847 | -0.4469 | Yes | ||
| 139 | E2F7 | 19884 | 17290 | -0.854 | -0.4374 | Yes | ||
| 140 | FBXL20 | 13010 | 17361 | -0.913 | -0.4306 | Yes | ||
| 141 | RAB11B | 9676 | 17414 | -0.948 | -0.4225 | Yes | ||
| 142 | TIAM1 | 22547 | 17464 | -0.988 | -0.4137 | Yes | ||
| 143 | KNTC1 | 16705 | 17471 | -0.992 | -0.4026 | Yes | ||
| 144 | BRMS1L | 21268 | 17494 | -1.001 | -0.3922 | Yes | ||
| 145 | USP52 | 3397 19841 | 17522 | -1.014 | -0.3819 | Yes | ||
| 146 | E2F1 | 14384 | 17550 | -1.032 | -0.3715 | Yes | ||
| 147 | NUTF2 | 12632 7529 | 17582 | -1.059 | -0.3609 | Yes | ||
| 148 | CDCA7 | 12437 | 17586 | -1.062 | -0.3488 | Yes | ||
| 149 | CDC25A | 8721 | 17619 | -1.092 | -0.3379 | Yes | ||
| 150 | RPS20 | 7438 | 17622 | -1.094 | -0.3254 | Yes | ||
| 151 | UNG | 10257 16744 | 17637 | -1.106 | -0.3133 | Yes | ||
| 152 | PPRC1 | 23808 | 17676 | -1.160 | -0.3020 | Yes | ||
| 153 | NASP | 2383 2387 6955 | 17824 | -1.310 | -0.2948 | Yes | ||
| 154 | HTF9C | 4886 1678 | 17954 | -1.450 | -0.2850 | Yes | ||
| 155 | APRIN | 4145 | 17968 | -1.465 | -0.2688 | Yes | ||
| 156 | ANP32A | 8592 4387 19415 | 18056 | -1.579 | -0.2552 | Yes | ||
| 157 | DPYSL2 | 3122 4561 8787 | 18061 | -1.580 | -0.2372 | Yes | ||
| 158 | FUS | 6135 | 18152 | -1.696 | -0.2225 | Yes | ||
| 159 | EIF5A | 11345 20379 6590 | 18169 | -1.721 | -0.2034 | Yes | ||
| 160 | POLA2 | 23988 | 18327 | -2.076 | -0.1879 | Yes | ||
| 161 | RPA2 | 2330 16057 | 18374 | -2.174 | -0.1653 | Yes | ||
| 162 | DNMT1 | 19217 | 18396 | -2.214 | -0.1408 | Yes | ||
| 163 | PRKCSH | 19535 | 18426 | -2.304 | -0.1158 | Yes | ||
| 164 | TOP1 | 5790 5789 10210 | 18440 | -2.355 | -0.0892 | Yes | ||
| 165 | FKBP5 | 23051 | 18492 | -2.579 | -0.0622 | Yes | ||
| 166 | POLD1 | 17847 | 18525 | -2.830 | -0.0312 | Yes | ||
| 167 | RFC1 | 16527 | 18551 | -3.119 | 0.0035 | Yes |