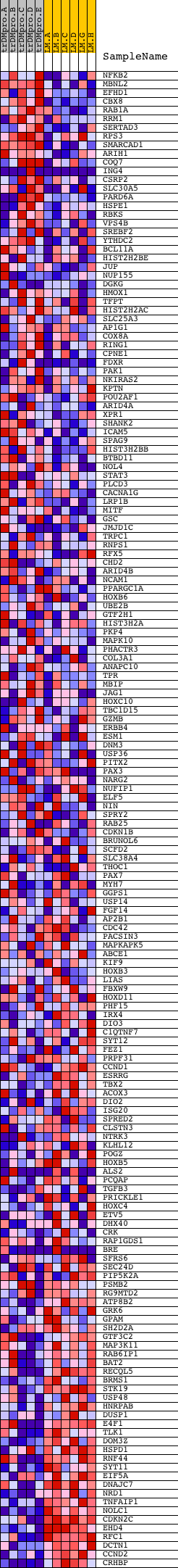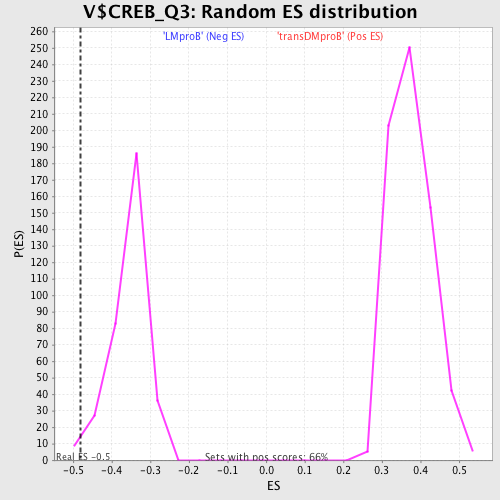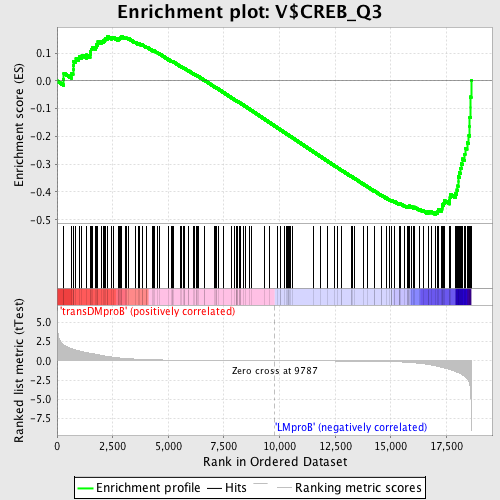
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | V$CREB_Q3 |
| Enrichment Score (ES) | -0.48102567 |
| Normalized Enrichment Score (NES) | -1.3569669 |
| Nominal p-value | 0.017595308 |
| FDR q-value | 0.5148636 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NFKB2 | 23810 | 286 | 2.058 | 0.0070 | No | ||
| 2 | MBNL2 | 21932 | 305 | 2.027 | 0.0281 | No | ||
| 3 | EFHD1 | 8267 | 638 | 1.596 | 0.0276 | No | ||
| 4 | CBX8 | 11401 | 716 | 1.513 | 0.0399 | No | ||
| 5 | RAB1A | 20942 | 724 | 1.502 | 0.0559 | No | ||
| 6 | RRM1 | 18163 | 740 | 1.487 | 0.0713 | No | ||
| 7 | SERTAD3 | 18327 | 848 | 1.388 | 0.0807 | No | ||
| 8 | RPS3 | 6549 11295 | 983 | 1.282 | 0.0874 | No | ||
| 9 | SMARCAD1 | 17427 | 1117 | 1.187 | 0.0932 | No | ||
| 10 | ARIH1 | 6210 | 1320 | 1.047 | 0.0937 | No | ||
| 11 | COQ7 | 17664 | 1480 | 0.978 | 0.0957 | No | ||
| 12 | ING4 | 6602 1155 1102 11371 | 1499 | 0.972 | 0.1054 | No | ||
| 13 | CSRP2 | 19883 | 1523 | 0.958 | 0.1146 | No | ||
| 14 | SLC30A5 | 3190 21361 | 1575 | 0.933 | 0.1220 | No | ||
| 15 | PARD6A | 12142 | 1745 | 0.832 | 0.1219 | No | ||
| 16 | HSPE1 | 9133 | 1756 | 0.828 | 0.1304 | No | ||
| 17 | RBKS | 16568 | 1834 | 0.783 | 0.1348 | No | ||
| 18 | VPS4B | 5446 | 1835 | 0.783 | 0.1433 | No | ||
| 19 | SREBF2 | 5509 | 1981 | 0.698 | 0.1431 | No | ||
| 20 | YTHDC2 | 23564 | 2076 | 0.653 | 0.1451 | No | ||
| 21 | BCL11A | 4691 | 2149 | 0.616 | 0.1480 | No | ||
| 22 | HIST2H2BE | 6651 | 2162 | 0.608 | 0.1539 | No | ||
| 23 | JUP | 4937 | 2256 | 0.557 | 0.1550 | No | ||
| 24 | NUP155 | 2298 5027 | 2275 | 0.552 | 0.1600 | No | ||
| 25 | DGKG | 4282 | 2448 | 0.488 | 0.1560 | No | ||
| 26 | HMOX1 | 18565 | 2520 | 0.460 | 0.1572 | No | ||
| 27 | TFPT | 1180 17993 | 2743 | 0.381 | 0.1493 | No | ||
| 28 | HIST2H2AC | 6650 | 2745 | 0.380 | 0.1534 | No | ||
| 29 | SLC25A3 | 19644 | 2818 | 0.359 | 0.1534 | No | ||
| 30 | AP1G1 | 4392 | 2860 | 0.348 | 0.1550 | No | ||
| 31 | COX8A | 8777 | 2879 | 0.344 | 0.1578 | No | ||
| 32 | RING1 | 23023 | 2882 | 0.343 | 0.1614 | No | ||
| 33 | CPNE1 | 11186 | 3068 | 0.295 | 0.1546 | No | ||
| 34 | FDXR | 1451 20154 | 3122 | 0.282 | 0.1548 | No | ||
| 35 | PAK1 | 9527 | 3203 | 0.265 | 0.1534 | No | ||
| 36 | NKIRAS2 | 12979 | 3505 | 0.213 | 0.1394 | No | ||
| 37 | KPTN | 18385 | 3669 | 0.191 | 0.1326 | No | ||
| 38 | POU2AF1 | 19455 | 3681 | 0.189 | 0.1341 | No | ||
| 39 | ARID4A | 2080 6215 | 3819 | 0.173 | 0.1286 | No | ||
| 40 | XPR1 | 5383 | 3827 | 0.172 | 0.1301 | No | ||
| 41 | SHANK2 | 17998 | 4017 | 0.150 | 0.1215 | No | ||
| 42 | ICAM5 | 19543 | 4037 | 0.148 | 0.1220 | No | ||
| 43 | SPAG9 | 1322 12904 7707 | 4304 | 0.123 | 0.1090 | No | ||
| 44 | HIST3H2BB | 11796 | 4334 | 0.121 | 0.1087 | No | ||
| 45 | BTBD11 | 13147 | 4366 | 0.119 | 0.1083 | No | ||
| 46 | NOL4 | 2028 11432 | 4495 | 0.112 | 0.1026 | No | ||
| 47 | STAT3 | 5525 9906 | 4604 | 0.106 | 0.0979 | No | ||
| 48 | PLCD3 | 20200 | 5028 | 0.084 | 0.0759 | No | ||
| 49 | CACNA1G | 1237 940 20292 | 5162 | 0.079 | 0.0696 | No | ||
| 50 | LRP1B | 8249 | 5183 | 0.078 | 0.0693 | No | ||
| 51 | MITF | 17349 | 5192 | 0.078 | 0.0698 | No | ||
| 52 | GSC | 20990 | 5224 | 0.077 | 0.0689 | No | ||
| 53 | JMJD1C | 11531 19996 | 5556 | 0.066 | 0.0517 | No | ||
| 54 | TRPC1 | 2984 19039 | 5606 | 0.065 | 0.0497 | No | ||
| 55 | RNPS1 | 9730 23361 | 5702 | 0.062 | 0.0453 | No | ||
| 56 | RFX5 | 7018 | 5714 | 0.062 | 0.0453 | No | ||
| 57 | CHD2 | 10847 | 5731 | 0.061 | 0.0451 | No | ||
| 58 | ARID4B | 21705 3233 | 5891 | 0.057 | 0.0371 | No | ||
| 59 | NCAM1 | 5149 | 5927 | 0.056 | 0.0359 | No | ||
| 60 | PPARGC1A | 16533 | 6137 | 0.051 | 0.0251 | No | ||
| 61 | HOXB6 | 20688 | 6165 | 0.050 | 0.0242 | No | ||
| 62 | UBE2B | 10245 | 6243 | 0.048 | 0.0205 | No | ||
| 63 | GTF2H1 | 4069 18236 | 6279 | 0.047 | 0.0191 | No | ||
| 64 | HIST3H2A | 6647 | 6325 | 0.046 | 0.0172 | No | ||
| 65 | PKP4 | 2822 15008 | 6337 | 0.046 | 0.0171 | No | ||
| 66 | MAPK10 | 11169 | 6603 | 0.041 | 0.0032 | No | ||
| 67 | PHACTR3 | 7900 2770 | 6619 | 0.041 | 0.0028 | No | ||
| 68 | COL3A1 | 14253 8765 | 7070 | 0.033 | -0.0212 | No | ||
| 69 | ANAPC10 | 7595 12760 7596 | 7106 | 0.033 | -0.0227 | No | ||
| 70 | TPR | 927 4255 | 7160 | 0.032 | -0.0253 | No | ||
| 71 | MBIP | 21063 2072 | 7162 | 0.032 | -0.0250 | No | ||
| 72 | JAG1 | 14415 | 7242 | 0.031 | -0.0289 | No | ||
| 73 | HOXC10 | 22342 | 7463 | 0.027 | -0.0406 | No | ||
| 74 | TBC1D15 | 19625 | 7842 | 0.022 | -0.0608 | No | ||
| 75 | GZMB | 21810 | 7955 | 0.020 | -0.0667 | No | ||
| 76 | ERBB4 | 13933 4678 | 8043 | 0.019 | -0.0712 | No | ||
| 77 | ESM1 | 21560 | 8050 | 0.019 | -0.0713 | No | ||
| 78 | DNM3 | 5948 | 8069 | 0.019 | -0.0721 | No | ||
| 79 | USP36 | 4648 8871 7804 | 8118 | 0.018 | -0.0745 | No | ||
| 80 | PITX2 | 15424 1878 | 8119 | 0.018 | -0.0743 | No | ||
| 81 | PAX3 | 13910 | 8195 | 0.017 | -0.0781 | No | ||
| 82 | NARG2 | 19390 | 8210 | 0.017 | -0.0787 | No | ||
| 83 | NUFIP1 | 21955 | 8250 | 0.017 | -0.0806 | No | ||
| 84 | ELF5 | 14936 2913 | 8251 | 0.017 | -0.0805 | No | ||
| 85 | NIN | 9463 2103 | 8365 | 0.016 | -0.0864 | No | ||
| 86 | SPRY2 | 21725 | 8458 | 0.014 | -0.0912 | No | ||
| 87 | RAB25 | 15287 | 8642 | 0.012 | -0.1010 | No | ||
| 88 | CDKN1B | 4512 8730 | 8749 | 0.011 | -0.1067 | No | ||
| 89 | BRUNOL6 | 19421 | 9303 | 0.005 | -0.1366 | No | ||
| 90 | SCFD2 | 5607 | 9549 | 0.003 | -0.1498 | No | ||
| 91 | SLC38A4 | 7614 12793 | 9894 | -0.001 | -0.1685 | No | ||
| 92 | THOC1 | 23623 | 10034 | -0.003 | -0.1760 | No | ||
| 93 | PAX7 | 15697 | 10053 | -0.003 | -0.1769 | No | ||
| 94 | MYH7 | 21828 4709 | 10207 | -0.005 | -0.1852 | No | ||
| 95 | GGPS1 | 3194 4776 | 10304 | -0.006 | -0.1903 | No | ||
| 96 | USP14 | 2001 23492 | 10335 | -0.006 | -0.1919 | No | ||
| 97 | FGF14 | 21716 3005 3463 | 10382 | -0.007 | -0.1943 | No | ||
| 98 | AP2B1 | 12958 7752 12957 1280 | 10405 | -0.007 | -0.1954 | No | ||
| 99 | CDC42 | 4503 8722 4504 2465 | 10448 | -0.007 | -0.1976 | No | ||
| 100 | PACSIN3 | 14948 8149 2776 2665 | 10478 | -0.008 | -0.1991 | No | ||
| 101 | MAPKAPK5 | 16387 | 10569 | -0.008 | -0.2039 | No | ||
| 102 | ABCE1 | 6270 | 11525 | -0.019 | -0.2554 | No | ||
| 103 | KIF9 | 19292 | 11843 | -0.023 | -0.2724 | No | ||
| 104 | HOXB3 | 20685 | 12141 | -0.027 | -0.2882 | No | ||
| 105 | LIAS | 13359 3620 13465 | 12451 | -0.031 | -0.3046 | No | ||
| 106 | FBXW9 | 18540 | 12605 | -0.034 | -0.3125 | No | ||
| 107 | HOXD11 | 9112 | 12772 | -0.037 | -0.3211 | No | ||
| 108 | PHF15 | 20467 8050 | 12804 | -0.037 | -0.3224 | No | ||
| 109 | IRX4 | 11944 | 13212 | -0.045 | -0.3440 | No | ||
| 110 | DIO3 | 19359 | 13221 | -0.045 | -0.3439 | No | ||
| 111 | C1QTNF7 | 16852 | 13261 | -0.046 | -0.3455 | No | ||
| 112 | SYT12 | 23962 | 13356 | -0.047 | -0.3501 | No | ||
| 113 | FEZ1 | 19520 3100 | 13754 | -0.057 | -0.3710 | No | ||
| 114 | PRPF31 | 7594 | 13955 | -0.063 | -0.3812 | No | ||
| 115 | CCND1 | 4487 4488 8707 17535 | 14255 | -0.072 | -0.3966 | No | ||
| 116 | ESRRG | 6447 | 14257 | -0.072 | -0.3959 | No | ||
| 117 | TBX2 | 20720 | 14571 | -0.084 | -0.4119 | No | ||
| 118 | ACOX3 | 3612 16867 | 14599 | -0.086 | -0.4124 | No | ||
| 119 | DIO2 | 21014 2159 | 14783 | -0.095 | -0.4213 | No | ||
| 120 | ISG20 | 18210 | 14959 | -0.107 | -0.4296 | No | ||
| 121 | SPRED2 | 8539 4332 | 15016 | -0.112 | -0.4314 | No | ||
| 122 | CLSTN3 | 17006 | 15036 | -0.113 | -0.4312 | No | ||
| 123 | NTRK3 | 2069 3652 3705 9492 | 15039 | -0.114 | -0.4301 | No | ||
| 124 | KLHL12 | 14125 | 15159 | -0.123 | -0.4352 | No | ||
| 125 | POGZ | 6031 1911 | 15165 | -0.124 | -0.4341 | No | ||
| 126 | HOXB5 | 20687 | 15386 | -0.145 | -0.4445 | No | ||
| 127 | ALS2 | 7882 13151 | 15397 | -0.147 | -0.4434 | No | ||
| 128 | PCQAP | 1661 13581 | 15405 | -0.148 | -0.4422 | No | ||
| 129 | TGFB3 | 10161 | 15438 | -0.151 | -0.4423 | No | ||
| 130 | PRICKLE1 | 8379 | 15622 | -0.176 | -0.4503 | No | ||
| 131 | HOXC4 | 4864 | 15741 | -0.198 | -0.4545 | No | ||
| 132 | ETV5 | 22630 | 15766 | -0.203 | -0.4536 | No | ||
| 133 | DHX40 | 20307 | 15812 | -0.211 | -0.4537 | No | ||
| 134 | CRK | 4559 1249 | 15815 | -0.212 | -0.4515 | No | ||
| 135 | RAP1GDS1 | 10487 | 15831 | -0.215 | -0.4500 | No | ||
| 136 | BRE | 3640 4226 16879 | 15947 | -0.240 | -0.4536 | No | ||
| 137 | SFRS6 | 14751 | 16039 | -0.262 | -0.4556 | No | ||
| 138 | SEC24D | 15429 | 16064 | -0.269 | -0.4540 | No | ||
| 139 | PIP5K2A | 14676 | 16304 | -0.334 | -0.4633 | No | ||
| 140 | PSMB2 | 2324 16078 | 16450 | -0.389 | -0.4669 | No | ||
| 141 | RG9MTD2 | 8443 | 16677 | -0.474 | -0.4740 | Yes | ||
| 142 | ATP8B2 | 12055 | 16678 | -0.474 | -0.4688 | Yes | ||
| 143 | GRK6 | 6449 | 16806 | -0.538 | -0.4699 | Yes | ||
| 144 | GPAM | 4795 | 17013 | -0.653 | -0.4739 | Yes | ||
| 145 | SH2D2A | 6569 | 17112 | -0.715 | -0.4714 | Yes | ||
| 146 | GTF3C2 | 7749 | 17120 | -0.721 | -0.4639 | Yes | ||
| 147 | MAP3K11 | 11163 | 17263 | -0.835 | -0.4625 | Yes | ||
| 148 | RAB6IP1 | 17676 | 17301 | -0.866 | -0.4550 | Yes | ||
| 149 | BAT2 | 23006 | 17305 | -0.867 | -0.4457 | Yes | ||
| 150 | RECQL5 | 20148 | 17382 | -0.922 | -0.4398 | Yes | ||
| 151 | BRMS1 | 8403 | 17409 | -0.946 | -0.4309 | Yes | ||
| 152 | STK19 | 23016 1569 1508 | 17639 | -1.108 | -0.4312 | Yes | ||
| 153 | USP48 | 16022 | 17658 | -1.135 | -0.4198 | Yes | ||
| 154 | HNRPAB | 9104 | 17686 | -1.173 | -0.4084 | Yes | ||
| 155 | DUSP1 | 23061 | 17895 | -1.371 | -0.4047 | Yes | ||
| 156 | E4F1 | 8875 | 17972 | -1.468 | -0.3928 | Yes | ||
| 157 | TLK1 | 14563 | 18014 | -1.532 | -0.3783 | Yes | ||
| 158 | DOM3Z | 1615 23271 | 18024 | -1.545 | -0.3619 | Yes | ||
| 159 | HSPD1 | 4078 9129 | 18026 | -1.548 | -0.3451 | Yes | ||
| 160 | RNF44 | 8364 | 18076 | -1.593 | -0.3303 | Yes | ||
| 161 | SYT11 | 15285 | 18127 | -1.662 | -0.3149 | Yes | ||
| 162 | EIF5A | 11345 20379 6590 | 18169 | -1.721 | -0.2983 | Yes | ||
| 163 | DNAJC7 | 20227 | 18213 | -1.809 | -0.2809 | Yes | ||
| 164 | NRD1 | 16140 | 18323 | -2.068 | -0.2642 | Yes | ||
| 165 | TNFAIP1 | 20333 | 18337 | -2.102 | -0.2420 | Yes | ||
| 166 | NOLC1 | 7704 | 18425 | -2.298 | -0.2216 | Yes | ||
| 167 | CDKN2C | 15806 2321 | 18509 | -2.692 | -0.1967 | Yes | ||
| 168 | EHD4 | 8275 | 18545 | -3.095 | -0.1648 | Yes | ||
| 169 | RFC1 | 16527 | 18551 | -3.119 | -0.1310 | Yes | ||
| 170 | DCTN1 | 1100 17392 | 18572 | -3.406 | -0.0949 | Yes | ||
| 171 | CCND2 | 16987 | 18573 | -3.414 | -0.0577 | Yes | ||
| 172 | CRHBP | 8785 | 18607 | -5.489 | 0.0005 | Yes |