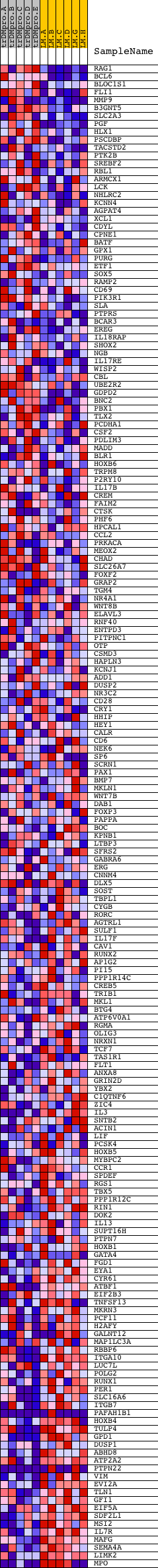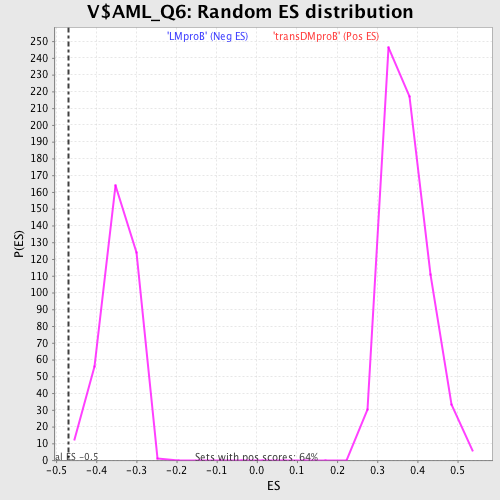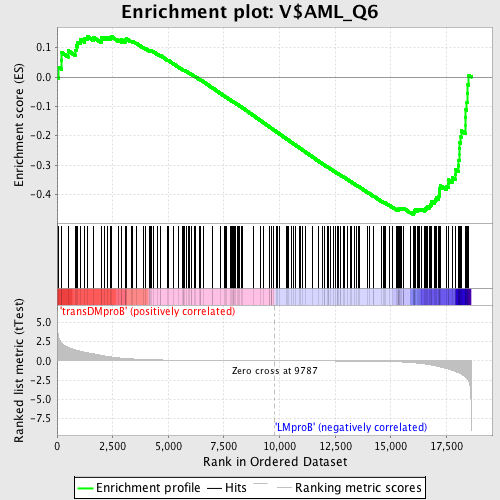
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | V$AML_Q6 |
| Enrichment Score (ES) | -0.46856114 |
| Normalized Enrichment Score (NES) | -1.356913 |
| Nominal p-value | 0.0028011205 |
| FDR q-value | 0.48828417 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RAG1 | 9689 | 78 | 2.899 | 0.0330 | No | ||
| 2 | BCL6 | 22624 | 205 | 2.250 | 0.0550 | No | ||
| 3 | BLOC1S1 | 3441 19586 | 213 | 2.228 | 0.0833 | No | ||
| 4 | FLI1 | 4729 | 493 | 1.724 | 0.0902 | No | ||
| 5 | MMP9 | 14732 | 834 | 1.402 | 0.0898 | No | ||
| 6 | B3GNT5 | 22824 | 854 | 1.385 | 0.1065 | No | ||
| 7 | SLC2A3 | 17010 | 933 | 1.314 | 0.1192 | No | ||
| 8 | PGF | 21020 | 1054 | 1.230 | 0.1284 | No | ||
| 9 | HLX1 | 9093 | 1243 | 1.101 | 0.1324 | No | ||
| 10 | PSCDBP | 10442 | 1356 | 1.024 | 0.1394 | No | ||
| 11 | TACSTD2 | 17126 | 1648 | 0.892 | 0.1351 | No | ||
| 12 | PTK2B | 21776 | 1979 | 0.701 | 0.1262 | No | ||
| 13 | SREBF2 | 5509 | 1981 | 0.698 | 0.1351 | No | ||
| 14 | RBL1 | 14373 | 2123 | 0.632 | 0.1355 | No | ||
| 15 | ARMCX1 | 24253 | 2261 | 0.556 | 0.1352 | No | ||
| 16 | LCK | 15746 | 2404 | 0.501 | 0.1340 | No | ||
| 17 | NHLRC2 | 23828 | 2445 | 0.488 | 0.1381 | No | ||
| 18 | KCNN4 | 18353 | 2754 | 0.378 | 0.1262 | No | ||
| 19 | AGPAT4 | 23384 | 2890 | 0.341 | 0.1233 | No | ||
| 20 | XCL1 | 13781 | 2899 | 0.339 | 0.1272 | No | ||
| 21 | CDYL | 3195 4513 3287 | 3057 | 0.297 | 0.1225 | No | ||
| 22 | CPNE1 | 11186 | 3068 | 0.295 | 0.1257 | No | ||
| 23 | BATF | 21200 | 3070 | 0.293 | 0.1294 | No | ||
| 24 | GPX1 | 19310 | 3096 | 0.286 | 0.1317 | No | ||
| 25 | PURG | 7943 | 3324 | 0.242 | 0.1225 | No | ||
| 26 | ETF1 | 23467 | 3389 | 0.232 | 0.1220 | No | ||
| 27 | SOX5 | 9850 1044 5480 16937 | 3554 | 0.205 | 0.1158 | No | ||
| 28 | RAMP2 | 20660 | 3901 | 0.161 | 0.0991 | No | ||
| 29 | CD69 | 16979 8717 | 3986 | 0.153 | 0.0965 | No | ||
| 30 | PIK3R1 | 3170 | 4146 | 0.138 | 0.0896 | No | ||
| 31 | SLA | 5449 | 4148 | 0.137 | 0.0913 | No | ||
| 32 | PTPRS | 1507 1565 1602 22928 | 4198 | 0.132 | 0.0904 | No | ||
| 33 | BCAR3 | 15432 | 4253 | 0.127 | 0.0891 | No | ||
| 34 | EREG | 4679 16797 | 4331 | 0.121 | 0.0864 | No | ||
| 35 | IL18RAP | 14262 | 4523 | 0.110 | 0.0775 | No | ||
| 36 | SHOX2 | 1802 5432 | 4632 | 0.104 | 0.0730 | No | ||
| 37 | NGB | 21016 | 4663 | 0.102 | 0.0726 | No | ||
| 38 | IL17RE | 434 17327 433 1049 | 4963 | 0.087 | 0.0575 | No | ||
| 39 | WISP2 | 14745 | 4986 | 0.086 | 0.0574 | No | ||
| 40 | CBL | 19154 | 5233 | 0.076 | 0.0451 | No | ||
| 41 | UBE2R2 | 16240 | 5466 | 0.069 | 0.0334 | No | ||
| 42 | GDPD2 | 24280 | 5623 | 0.064 | 0.0257 | No | ||
| 43 | BNC2 | 15850 | 5676 | 0.063 | 0.0237 | No | ||
| 44 | PBX1 | 5224 | 5690 | 0.062 | 0.0238 | No | ||
| 45 | TLX2 | 5773 1165 | 5734 | 0.061 | 0.0223 | No | ||
| 46 | PCDHA1 | 8571 4368 | 5794 | 0.060 | 0.0198 | No | ||
| 47 | CSF2 | 20454 | 5887 | 0.057 | 0.0156 | No | ||
| 48 | PDLIM3 | 18625 | 5955 | 0.055 | 0.0126 | No | ||
| 49 | MADD | 2926 14526 | 6035 | 0.053 | 0.0090 | No | ||
| 50 | BLR1 | 19145 3154 | 6050 | 0.053 | 0.0090 | No | ||
| 51 | HOXB6 | 20688 | 6165 | 0.050 | 0.0034 | No | ||
| 52 | TRPM8 | 9363 | 6216 | 0.049 | 0.0013 | No | ||
| 53 | P2RY10 | 2613 24269 | 6420 | 0.045 | -0.0091 | No | ||
| 54 | IL17B | 12074 | 6442 | 0.044 | -0.0097 | No | ||
| 55 | CREM | 2005 1965 23496 1951 | 6453 | 0.044 | -0.0097 | No | ||
| 56 | FAIM2 | 13027 | 6573 | 0.042 | -0.0156 | No | ||
| 57 | CTSK | 15506 | 6968 | 0.035 | -0.0365 | No | ||
| 58 | PHF6 | 2648 24340 | 6970 | 0.035 | -0.0361 | No | ||
| 59 | HPCAL1 | 21307 2123 | 7361 | 0.029 | -0.0569 | No | ||
| 60 | CCL2 | 9788 | 7519 | 0.026 | -0.0651 | No | ||
| 61 | PRKACA | 18549 3844 | 7558 | 0.026 | -0.0668 | No | ||
| 62 | MEOX2 | 21285 | 7635 | 0.025 | -0.0706 | No | ||
| 63 | CHAD | 20702 | 7781 | 0.023 | -0.0782 | No | ||
| 64 | SLC26A7 | 5538 2372 | 7828 | 0.022 | -0.0804 | No | ||
| 65 | FOXF2 | 21677 | 7897 | 0.021 | -0.0838 | No | ||
| 66 | GRAP2 | 5113 9398 | 7924 | 0.021 | -0.0850 | No | ||
| 67 | TGM4 | 11662 | 7947 | 0.020 | -0.0859 | No | ||
| 68 | NR4A1 | 9099 | 7970 | 0.020 | -0.0869 | No | ||
| 69 | WNT8B | 23841 | 8034 | 0.019 | -0.0900 | No | ||
| 70 | ELAVL3 | 9136 | 8116 | 0.018 | -0.0942 | No | ||
| 71 | RNF40 | 6133 | 8150 | 0.018 | -0.0958 | No | ||
| 72 | ENTPD3 | 19266 | 8175 | 0.018 | -0.0968 | No | ||
| 73 | PITPNC1 | 7759 | 8278 | 0.017 | -0.1022 | No | ||
| 74 | OTP | 21582 | 8333 | 0.016 | -0.1049 | No | ||
| 75 | CSMD3 | 10735 7938 | 8823 | 0.010 | -0.1313 | No | ||
| 76 | HAPLN3 | 17792 | 9135 | 0.007 | -0.1481 | No | ||
| 77 | KCNJ1 | 3121 12110 | 9272 | 0.006 | -0.1554 | No | ||
| 78 | ADD1 | 3615 4350 3590 | 9526 | 0.003 | -0.1691 | No | ||
| 79 | DUSP2 | 14865 | 9559 | 0.003 | -0.1708 | No | ||
| 80 | NR3C2 | 18562 13798 | 9628 | 0.002 | -0.1744 | No | ||
| 81 | CD28 | 14239 4092 | 9636 | 0.002 | -0.1748 | No | ||
| 82 | CRY1 | 19662 | 9653 | 0.002 | -0.1757 | No | ||
| 83 | HHIP | 4849 | 9733 | 0.001 | -0.1799 | No | ||
| 84 | HEY1 | 15378 | 9852 | -0.001 | -0.1863 | No | ||
| 85 | CALR | 18814 | 9917 | -0.002 | -0.1898 | No | ||
| 86 | CD6 | 3669 23740 | 10003 | -0.003 | -0.1944 | No | ||
| 87 | NEK6 | 15018 | 10324 | -0.006 | -0.2116 | No | ||
| 88 | SP6 | 8178 13678 13679 | 10366 | -0.006 | -0.2138 | No | ||
| 89 | SCRN1 | 7649 12840 | 10418 | -0.007 | -0.2165 | No | ||
| 90 | PAX1 | 9529 | 10521 | -0.008 | -0.2219 | No | ||
| 91 | BMP7 | 14324 | 10622 | -0.009 | -0.2272 | No | ||
| 92 | MKLN1 | 6584 11339 | 10695 | -0.010 | -0.2310 | No | ||
| 93 | WNT7B | 22172 | 10872 | -0.012 | -0.2404 | No | ||
| 94 | DAB1 | 4594 8834 | 10940 | -0.012 | -0.2439 | No | ||
| 95 | FOXP3 | 9804 5426 | 11007 | -0.013 | -0.2473 | No | ||
| 96 | PAPPA | 16190 | 11171 | -0.015 | -0.2559 | No | ||
| 97 | BOC | 22587 | 11459 | -0.018 | -0.2713 | No | ||
| 98 | KPNB1 | 20274 | 11745 | -0.022 | -0.2865 | No | ||
| 99 | LTBP3 | 5008 23785 9288 | 11938 | -0.024 | -0.2966 | No | ||
| 100 | SFRS2 | 9807 20136 | 12015 | -0.025 | -0.3004 | No | ||
| 101 | GABRA6 | 20491 | 12033 | -0.026 | -0.3010 | No | ||
| 102 | ERG | 1686 8915 | 12169 | -0.027 | -0.3079 | No | ||
| 103 | CNNM4 | 14273 | 12191 | -0.028 | -0.3087 | No | ||
| 104 | DLX5 | 17219 1058 | 12199 | -0.028 | -0.3087 | No | ||
| 105 | SOST | 20210 | 12301 | -0.029 | -0.3139 | No | ||
| 106 | TBPL1 | 19805 | 12418 | -0.031 | -0.3197 | No | ||
| 107 | CYGB | 20139 | 12513 | -0.032 | -0.3244 | No | ||
| 108 | RORC | 9732 | 12608 | -0.034 | -0.3291 | No | ||
| 109 | AGTRL1 | 14969 | 12611 | -0.034 | -0.3288 | No | ||
| 110 | SULF1 | 6285 | 12630 | -0.034 | -0.3293 | No | ||
| 111 | IL17F | 13992 | 12741 | -0.036 | -0.3348 | No | ||
| 112 | CAV1 | 8698 | 12742 | -0.036 | -0.3344 | No | ||
| 113 | RUNX2 | 4480 8700 | 12860 | -0.038 | -0.3402 | No | ||
| 114 | AP1G2 | 2677 21827 | 12929 | -0.039 | -0.3434 | No | ||
| 115 | PI15 | 13586 | 13051 | -0.042 | -0.3494 | No | ||
| 116 | PPP1R14C | 8006 8007 3363 | 13185 | -0.044 | -0.3561 | No | ||
| 117 | CREB5 | 10551 | 13249 | -0.045 | -0.3589 | No | ||
| 118 | TRIB1 | 22467 | 13385 | -0.048 | -0.3656 | No | ||
| 119 | MKL1 | 5864 | 13446 | -0.050 | -0.3682 | No | ||
| 120 | BTG4 | 19456 | 13564 | -0.052 | -0.3739 | No | ||
| 121 | ATP6V0A1 | 8640 4424 1197 | 13586 | -0.053 | -0.3744 | No | ||
| 122 | RGMA | 18213 | 13970 | -0.063 | -0.3944 | No | ||
| 123 | OLIG3 | 20084 | 14030 | -0.065 | -0.3967 | No | ||
| 124 | NRXN1 | 1581 1575 22875 | 14220 | -0.071 | -0.4061 | No | ||
| 125 | TCF7 | 1467 20466 | 14232 | -0.071 | -0.4058 | No | ||
| 126 | TAS1R1 | 15657 | 14586 | -0.085 | -0.4238 | No | ||
| 127 | FLT1 | 3483 16287 | 14684 | -0.090 | -0.4279 | No | ||
| 128 | ANXA8 | 22046 | 14712 | -0.091 | -0.4282 | No | ||
| 129 | GRIN2D | 9042 | 14738 | -0.093 | -0.4284 | No | ||
| 130 | YBX2 | 20818 | 14922 | -0.105 | -0.4370 | No | ||
| 131 | C1QTNF6 | 13063 | 15080 | -0.117 | -0.4440 | No | ||
| 132 | ZIC4 | 5992 | 15258 | -0.132 | -0.4519 | No | ||
| 133 | IL3 | 20453 | 15288 | -0.135 | -0.4517 | No | ||
| 134 | SNTB2 | 3887 18476 | 15342 | -0.141 | -0.4528 | No | ||
| 135 | ACIN1 | 3319 2807 21832 | 15351 | -0.141 | -0.4514 | No | ||
| 136 | LIF | 956 667 20960 | 15353 | -0.141 | -0.4497 | No | ||
| 137 | PCSK4 | 19696 3296 3394 18705 | 15375 | -0.144 | -0.4490 | No | ||
| 138 | HOXB5 | 20687 | 15386 | -0.145 | -0.4477 | No | ||
| 139 | MYBPC2 | 17848 | 15452 | -0.153 | -0.4492 | No | ||
| 140 | CCR1 | 18951 | 15473 | -0.155 | -0.4483 | No | ||
| 141 | SPDEF | 23055 | 15551 | -0.166 | -0.4504 | No | ||
| 142 | RGS1 | 3970 6936 | 15581 | -0.169 | -0.4498 | No | ||
| 143 | TBX5 | 3531 5634 | 15588 | -0.172 | -0.4479 | No | ||
| 144 | PPP1R12C | 6115 17987 | 15878 | -0.224 | -0.4607 | No | ||
| 145 | RIN1 | 10349 | 16024 | -0.258 | -0.4653 | Yes | ||
| 146 | DOK2 | 354 | 16030 | -0.259 | -0.4622 | Yes | ||
| 147 | IL13 | 20461 | 16056 | -0.266 | -0.4601 | Yes | ||
| 148 | SUPT16H | 8541 | 16057 | -0.266 | -0.4567 | Yes | ||
| 149 | PTPN7 | 11500 | 16073 | -0.271 | -0.4541 | Yes | ||
| 150 | HOXB1 | 20684 | 16102 | -0.277 | -0.4520 | Yes | ||
| 151 | GATA4 | 21792 4755 | 16200 | -0.306 | -0.4534 | Yes | ||
| 152 | FGD1 | 24228 | 16229 | -0.312 | -0.4509 | Yes | ||
| 153 | EYA1 | 4695 4061 | 16293 | -0.331 | -0.4501 | Yes | ||
| 154 | CYR61 | 9159 15145 9311 5024 | 16397 | -0.368 | -0.4509 | Yes | ||
| 155 | ATBF1 | 916 8633 | 16509 | -0.410 | -0.4517 | Yes | ||
| 156 | EIF2B3 | 16118 | 16557 | -0.426 | -0.4488 | Yes | ||
| 157 | TNFSF13 | 12806 | 16596 | -0.437 | -0.4452 | Yes | ||
| 158 | MKRN3 | 17810 | 16651 | -0.463 | -0.4422 | Yes | ||
| 159 | PCF11 | 121 | 16733 | -0.505 | -0.4401 | Yes | ||
| 160 | H2AFY | 21447 | 16783 | -0.527 | -0.4360 | Yes | ||
| 161 | GALNT12 | 16214 | 16824 | -0.544 | -0.4312 | Yes | ||
| 162 | MAP1LC3A | 14774 2741 | 16836 | -0.553 | -0.4247 | Yes | ||
| 163 | RBBP6 | 5365 | 16981 | -0.632 | -0.4244 | Yes | ||
| 164 | ITGA10 | 15493 | 17000 | -0.647 | -0.4171 | Yes | ||
| 165 | LUC7L | 47 1520 | 17070 | -0.694 | -0.4119 | Yes | ||
| 166 | POLG2 | 20180 | 17158 | -0.748 | -0.4070 | Yes | ||
| 167 | RUNX1 | 4481 | 17175 | -0.761 | -0.3981 | Yes | ||
| 168 | PER1 | 20827 | 17182 | -0.767 | -0.3886 | Yes | ||
| 169 | SLC16A6 | 4186 | 17201 | -0.783 | -0.3795 | Yes | ||
| 170 | ITGB7 | 22112 | 17234 | -0.814 | -0.3708 | Yes | ||
| 171 | PAFAH1B1 | 1340 5220 9524 | 17497 | -1.002 | -0.3722 | Yes | ||
| 172 | HOXB4 | 20686 | 17584 | -1.062 | -0.3632 | Yes | ||
| 173 | TULP4 | 12740 | 17591 | -1.066 | -0.3499 | Yes | ||
| 174 | GPD1 | 22361 | 17760 | -1.242 | -0.3430 | Yes | ||
| 175 | DUSP1 | 23061 | 17895 | -1.371 | -0.3327 | Yes | ||
| 176 | ABHD8 | 18847 | 17908 | -1.387 | -0.3156 | Yes | ||
| 177 | ATP2A2 | 4421 3481 | 18034 | -1.561 | -0.3023 | Yes | ||
| 178 | PTPN22 | 1775 15470 | 18055 | -1.578 | -0.2831 | Yes | ||
| 179 | VIM | 80 | 18085 | -1.606 | -0.2641 | Yes | ||
| 180 | EVI2A | 4690 | 18087 | -1.609 | -0.2435 | Yes | ||
| 181 | TLN1 | 15899 | 18102 | -1.621 | -0.2234 | Yes | ||
| 182 | GFI1 | 16451 | 18149 | -1.695 | -0.2042 | Yes | ||
| 183 | EIF5A | 11345 20379 6590 | 18169 | -1.721 | -0.1831 | Yes | ||
| 184 | SDF2L1 | 22649 | 18344 | -2.114 | -0.1654 | Yes | ||
| 185 | MSI2 | 8030 1268 | 18347 | -2.118 | -0.1384 | Yes | ||
| 186 | IL7R | 9175 4922 | 18371 | -2.168 | -0.1118 | Yes | ||
| 187 | MAFG | 5060 | 18420 | -2.276 | -0.0852 | Yes | ||
| 188 | SEMA4A | 15289 | 18434 | -2.325 | -0.0560 | Yes | ||
| 189 | LIMK2 | 9278 1296 | 18462 | -2.463 | -0.0259 | Yes | ||
| 190 | MPO | 5116 20710 1324 | 18506 | -2.663 | 0.0060 | Yes |