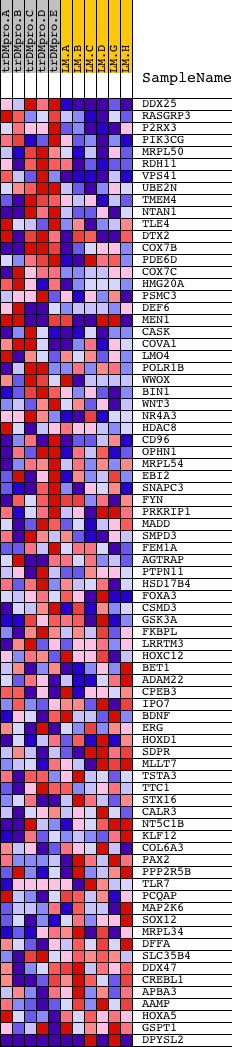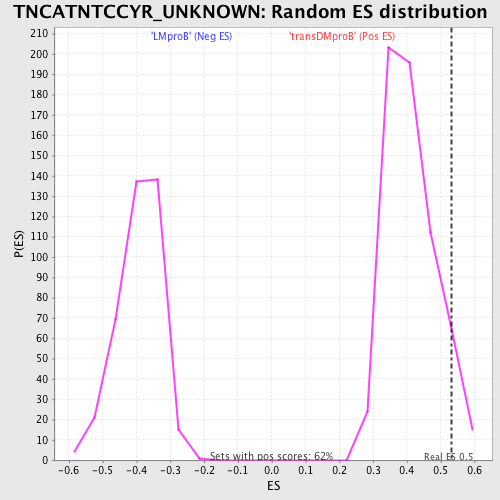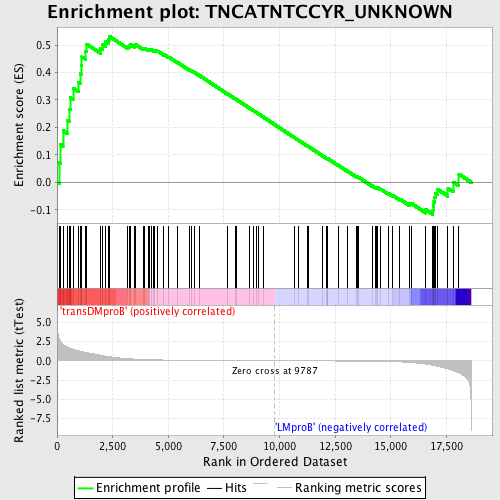
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | TNCATNTCCYR_UNKNOWN |
| Enrichment Score (ES) | 0.5326075 |
| Normalized Enrichment Score (NES) | 1.2999879 |
| Nominal p-value | 0.058536585 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DDX25 | 19184 | 90 | 2.792 | 0.0714 | Yes | ||
| 2 | RASGRP3 | 10767 | 134 | 2.549 | 0.1387 | Yes | ||
| 3 | P2RX3 | 10446 | 267 | 2.088 | 0.1886 | Yes | ||
| 4 | PIK3CG | 6635 | 476 | 1.747 | 0.2251 | Yes | ||
| 5 | MRPL50 | 11372 | 557 | 1.649 | 0.2658 | Yes | ||
| 6 | RDH11 | 21031 | 587 | 1.632 | 0.3088 | Yes | ||
| 7 | VPS41 | 3261 21701 | 718 | 1.512 | 0.3431 | Yes | ||
| 8 | UBE2N | 8216 19898 | 968 | 1.290 | 0.3649 | Yes | ||
| 9 | TMEM4 | 19842 | 1033 | 1.246 | 0.3955 | Yes | ||
| 10 | NTAN1 | 22852 | 1078 | 1.212 | 0.4262 | Yes | ||
| 11 | TLE4 | 23720 3697 | 1115 | 1.187 | 0.4567 | Yes | ||
| 12 | DTX2 | 16674 3488 | 1267 | 1.089 | 0.4783 | Yes | ||
| 13 | COX7B | 7260 | 1326 | 1.043 | 0.5036 | Yes | ||
| 14 | PDE6D | 13897 3945 | 1956 | 0.715 | 0.4892 | Yes | ||
| 15 | COX7C | 8776 | 2035 | 0.672 | 0.5034 | Yes | ||
| 16 | HMG20A | 19436 | 2181 | 0.596 | 0.5118 | Yes | ||
| 17 | PSMC3 | 9636 | 2313 | 0.538 | 0.5195 | Yes | ||
| 18 | DEF6 | 23319 | 2337 | 0.527 | 0.5326 | Yes | ||
| 19 | MEN1 | 3716 9382 | 3157 | 0.276 | 0.4960 | No | ||
| 20 | CASK | 24184 2618 2604 | 3264 | 0.254 | 0.4972 | No | ||
| 21 | COVA1 | 2636 24158 | 3297 | 0.246 | 0.5022 | No | ||
| 22 | LMO4 | 15151 | 3474 | 0.218 | 0.4986 | No | ||
| 23 | POLR1B | 14857 | 3509 | 0.213 | 0.5026 | No | ||
| 24 | WWOX | 13473 18458 18459 | 3874 | 0.165 | 0.4875 | No | ||
| 25 | BIN1 | 6634 2017 | 3936 | 0.157 | 0.4885 | No | ||
| 26 | WNT3 | 20635 | 4109 | 0.141 | 0.4831 | No | ||
| 27 | NR4A3 | 9473 16212 5183 | 4142 | 0.138 | 0.4851 | No | ||
| 28 | HDAC8 | 24090 | 4224 | 0.130 | 0.4843 | No | ||
| 29 | CD96 | 22580 | 4353 | 0.120 | 0.4807 | No | ||
| 30 | OPHN1 | 24102 | 4398 | 0.117 | 0.4815 | No | ||
| 31 | MRPL54 | 19678 | 4513 | 0.110 | 0.4784 | No | ||
| 32 | EBI2 | 21718 | 4802 | 0.094 | 0.4654 | No | ||
| 33 | SNAPC3 | 13397 2456 8085 | 5022 | 0.084 | 0.4559 | No | ||
| 34 | FYN | 3375 3395 20052 | 5417 | 0.071 | 0.4366 | No | ||
| 35 | PRKRIP1 | 12407 7336 | 5939 | 0.056 | 0.4100 | No | ||
| 36 | MADD | 2926 14526 | 6035 | 0.053 | 0.4063 | No | ||
| 37 | SMPD3 | 18757 | 6166 | 0.050 | 0.4007 | No | ||
| 38 | FEM1A | 23185 | 6421 | 0.045 | 0.3882 | No | ||
| 39 | AGTRAP | 15677 | 7646 | 0.025 | 0.3229 | No | ||
| 40 | PTPN11 | 5326 16391 9660 | 8010 | 0.020 | 0.3038 | No | ||
| 41 | HSD17B4 | 23560 | 8078 | 0.019 | 0.3007 | No | ||
| 42 | FOXA3 | 17949 | 8662 | 0.012 | 0.2696 | No | ||
| 43 | CSMD3 | 10735 7938 | 8823 | 0.010 | 0.2612 | No | ||
| 44 | GSK3A | 405 | 8943 | 0.009 | 0.2551 | No | ||
| 45 | FKBPL | 23273 | 9058 | 0.008 | 0.2491 | No | ||
| 46 | LRRTM3 | 19744 | 9292 | 0.005 | 0.2367 | No | ||
| 47 | HOXC12 | 22343 | 10663 | -0.009 | 0.1631 | No | ||
| 48 | BET1 | 17226 | 10847 | -0.011 | 0.1535 | No | ||
| 49 | ADAM22 | 8549 | 11264 | -0.016 | 0.1315 | No | ||
| 50 | CPEB3 | 5539 | 11302 | -0.016 | 0.1299 | No | ||
| 51 | IPO7 | 6130 | 11947 | -0.025 | 0.0959 | No | ||
| 52 | BDNF | 14926 2797 | 12118 | -0.027 | 0.0874 | No | ||
| 53 | ERG | 1686 8915 | 12169 | -0.027 | 0.0855 | No | ||
| 54 | HOXD1 | 14981 | 12172 | -0.027 | 0.0861 | No | ||
| 55 | SDPR | 14252 | 12660 | -0.035 | 0.0608 | No | ||
| 56 | MLLT7 | 24278 | 13035 | -0.042 | 0.0418 | No | ||
| 57 | TSTA3 | 22253 | 13438 | -0.049 | 0.0214 | No | ||
| 58 | TTC1 | 7342 | 13488 | -0.051 | 0.0202 | No | ||
| 59 | STX16 | 10464 | 13532 | -0.052 | 0.0193 | No | ||
| 60 | CALR3 | 13099 3791 3828 3909 | 14156 | -0.069 | -0.0125 | No | ||
| 61 | NT5C1B | 21315 | 14320 | -0.075 | -0.0192 | No | ||
| 62 | KLF12 | 4960 2637 21732 9228 | 14352 | -0.076 | -0.0188 | No | ||
| 63 | COL6A3 | 4546 13885 13884 | 14403 | -0.078 | -0.0194 | No | ||
| 64 | PAX2 | 9530 24416 | 14554 | -0.084 | -0.0252 | No | ||
| 65 | PPP2R5B | 23994 | 14893 | -0.102 | -0.0406 | No | ||
| 66 | TLR7 | 24004 | 15074 | -0.117 | -0.0472 | No | ||
| 67 | PCQAP | 1661 13581 | 15405 | -0.148 | -0.0609 | No | ||
| 68 | MAP2K6 | 20614 1414 | 15832 | -0.216 | -0.0780 | No | ||
| 69 | SOX12 | 5478 | 15908 | -0.229 | -0.0758 | No | ||
| 70 | MRPL34 | 18577 | 16575 | -0.430 | -0.1000 | No | ||
| 71 | DFFA | 4627 | 16893 | -0.585 | -0.1011 | No | ||
| 72 | SLC35B4 | 17187 | 16899 | -0.585 | -0.0854 | No | ||
| 73 | DDX47 | 17261 12577 1133 | 16937 | -0.602 | -0.0709 | No | ||
| 74 | CREBL1 | 23274 | 16974 | -0.630 | -0.0557 | No | ||
| 75 | APBA3 | 12180 | 16997 | -0.646 | -0.0392 | No | ||
| 76 | AAMP | 10406 | 17110 | -0.714 | -0.0258 | No | ||
| 77 | HOXA5 | 17149 | 17569 | -1.050 | -0.0218 | No | ||
| 78 | GSPT1 | 9046 | 17801 | -1.282 | 0.0008 | No | ||
| 79 | DPYSL2 | 3122 4561 8787 | 18061 | -1.580 | 0.0299 | No |