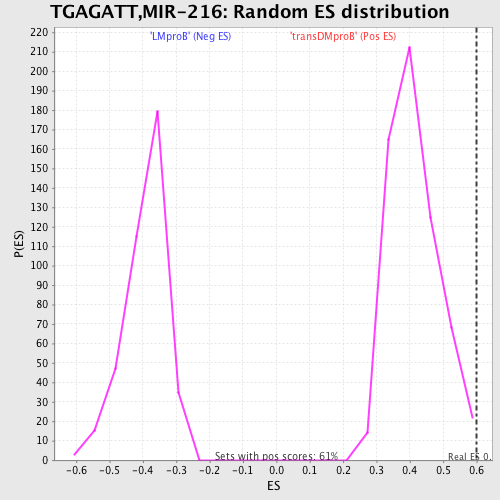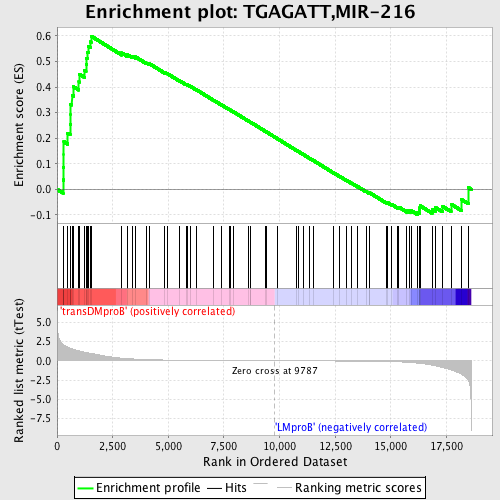
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
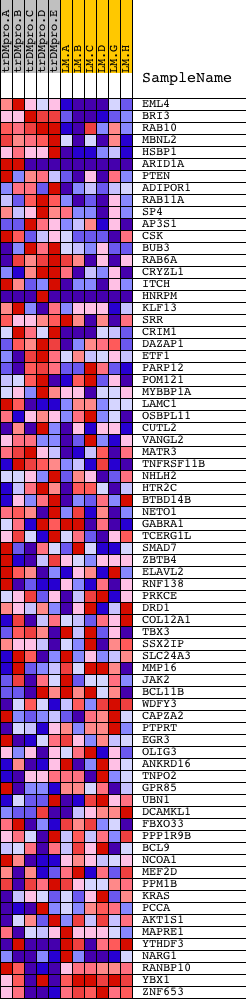
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | TGAGATT,MIR-216 |
| Enrichment Score (ES) | 0.5993121 |
| Normalized Enrichment Score (NES) | 1.4544393 |
| Nominal p-value | 0.011551155 |
| FDR q-value | 0.81661165 |
| FWER p-Value | 0.972 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EML4 | 8122 | 269 | 2.085 | 0.0369 | Yes | ||
| 2 | BRI3 | 15101 | 289 | 2.056 | 0.0866 | Yes | ||
| 3 | RAB10 | 2090 21119 | 303 | 2.030 | 0.1360 | Yes | ||
| 4 | MBNL2 | 21932 | 305 | 2.027 | 0.1860 | Yes | ||
| 5 | HSBP1 | 18451 | 477 | 1.746 | 0.2198 | Yes | ||
| 6 | ARID1A | 8214 2501 13554 | 586 | 1.633 | 0.2542 | Yes | ||
| 7 | PTEN | 5305 | 607 | 1.620 | 0.2931 | Yes | ||
| 8 | ADIPOR1 | 14126 | 612 | 1.616 | 0.3328 | Yes | ||
| 9 | RAB11A | 7013 | 669 | 1.562 | 0.3683 | Yes | ||
| 10 | SP4 | 20972 | 738 | 1.491 | 0.4014 | Yes | ||
| 11 | AP3S1 | 8604 | 980 | 1.285 | 0.4201 | Yes | ||
| 12 | CSK | 8805 | 991 | 1.274 | 0.4510 | Yes | ||
| 13 | BUB3 | 18045 | 1222 | 1.118 | 0.4662 | Yes | ||
| 14 | RAB6A | 18173 3905 | 1300 | 1.066 | 0.4883 | Yes | ||
| 15 | CRYZL1 | 1644 12385 | 1324 | 1.045 | 0.5128 | Yes | ||
| 16 | ITCH | 4928 9185 | 1354 | 1.025 | 0.5365 | Yes | ||
| 17 | HNRPM | 1511 13370 | 1408 | 1.001 | 0.5584 | Yes | ||
| 18 | KLF13 | 17807 | 1511 | 0.964 | 0.5767 | Yes | ||
| 19 | SRR | 6566 11328 | 1529 | 0.956 | 0.5993 | Yes | ||
| 20 | CRIM1 | 403 | 2903 | 0.338 | 0.5336 | No | ||
| 21 | DAZAP1 | 19945 | 3160 | 0.275 | 0.5266 | No | ||
| 22 | ETF1 | 23467 | 3389 | 0.232 | 0.5200 | No | ||
| 23 | PARP12 | 6335 | 3519 | 0.211 | 0.5183 | No | ||
| 24 | POM121 | 16343 | 4034 | 0.149 | 0.4942 | No | ||
| 25 | MYBBP1A | 5217 1232 5216 | 4132 | 0.139 | 0.4924 | No | ||
| 26 | LAMC1 | 13805 | 4841 | 0.092 | 0.4565 | No | ||
| 27 | OSBPL11 | 22781 | 4939 | 0.088 | 0.4535 | No | ||
| 28 | CUTL2 | 13110 4578 | 5510 | 0.067 | 0.4244 | No | ||
| 29 | VANGL2 | 4109 8220 | 5796 | 0.060 | 0.4105 | No | ||
| 30 | MATR3 | 2043 5081 | 5845 | 0.058 | 0.4093 | No | ||
| 31 | TNFRSF11B | 22296 | 5986 | 0.054 | 0.4031 | No | ||
| 32 | NHLH2 | 9462 | 6244 | 0.048 | 0.3904 | No | ||
| 33 | HTR2C | 24230 | 7043 | 0.034 | 0.3482 | No | ||
| 34 | BTBD14B | 7344 7343 | 7386 | 0.028 | 0.3305 | No | ||
| 35 | NETO1 | 6372 1970 | 7759 | 0.023 | 0.3110 | No | ||
| 36 | GABRA1 | 20492 | 7778 | 0.023 | 0.3106 | No | ||
| 37 | TCERG1L | 7690 | 7930 | 0.021 | 0.3030 | No | ||
| 38 | SMAD7 | 23513 | 8609 | 0.013 | 0.2667 | No | ||
| 39 | ZBTB4 | 13267 | 8682 | 0.012 | 0.2631 | No | ||
| 40 | ELAVL2 | 15839 | 8685 | 0.012 | 0.2633 | No | ||
| 41 | RNF138 | 2006 23613 2029 2037 | 9381 | 0.004 | 0.2259 | No | ||
| 42 | PRKCE | 9575 | 9432 | 0.004 | 0.2233 | No | ||
| 43 | DRD1 | 4638 | 9892 | -0.001 | 0.1986 | No | ||
| 44 | COL12A1 | 3143 19054 | 10745 | -0.010 | 0.1529 | No | ||
| 45 | TBX3 | 16723 | 10868 | -0.012 | 0.1466 | No | ||
| 46 | SSX2IP | 13617 | 11059 | -0.014 | 0.1367 | No | ||
| 47 | SLC24A3 | 14818 | 11354 | -0.017 | 0.1213 | No | ||
| 48 | MMP16 | 5109 | 11545 | -0.019 | 0.1115 | No | ||
| 49 | JAK2 | 23893 9197 3706 | 12438 | -0.031 | 0.0641 | No | ||
| 50 | BCL11B | 7190 | 12709 | -0.035 | 0.0505 | No | ||
| 51 | WDFY3 | 7789 3607 16462 | 12992 | -0.041 | 0.0363 | No | ||
| 52 | CAPZA2 | 8688 991 | 13222 | -0.045 | 0.0250 | No | ||
| 53 | PTPRT | 5333 | 13519 | -0.051 | 0.0103 | No | ||
| 54 | EGR3 | 4656 | 13898 | -0.061 | -0.0086 | No | ||
| 55 | OLIG3 | 20084 | 14030 | -0.065 | -0.0140 | No | ||
| 56 | ANKRD16 | 6804 | 14048 | -0.065 | -0.0133 | No | ||
| 57 | TNPO2 | 5608 | 14797 | -0.096 | -0.0513 | No | ||
| 58 | GPR85 | 7227 1048 | 14861 | -0.100 | -0.0522 | No | ||
| 59 | UBN1 | 22867 | 15034 | -0.113 | -0.0587 | No | ||
| 60 | DCAMKL1 | 8838 4600 | 15313 | -0.138 | -0.0703 | No | ||
| 61 | FBXO33 | 528 | 15354 | -0.141 | -0.0690 | No | ||
| 62 | PPP1R9B | 20698 | 15722 | -0.194 | -0.0840 | No | ||
| 63 | BCL9 | 15232 | 15819 | -0.213 | -0.0839 | No | ||
| 64 | NCOA1 | 5154 | 15919 | -0.233 | -0.0835 | No | ||
| 65 | MEF2D | 9379 | 16193 | -0.303 | -0.0907 | No | ||
| 66 | PPM1B | 5280 1567 5281 | 16297 | -0.332 | -0.0881 | No | ||
| 67 | KRAS | 9247 | 16300 | -0.333 | -0.0800 | No | ||
| 68 | PCCA | 21926 | 16306 | -0.335 | -0.0720 | No | ||
| 69 | AKT1S1 | 12557 | 16312 | -0.338 | -0.0639 | No | ||
| 70 | MAPRE1 | 4652 | 16854 | -0.563 | -0.0792 | No | ||
| 71 | YTHDF3 | 15630 10469 6019 | 17016 | -0.654 | -0.0718 | No | ||
| 72 | NARG1 | 7934 917 | 17319 | -0.884 | -0.0663 | No | ||
| 73 | RANBP10 | 18764 | 17708 | -1.189 | -0.0578 | No | ||
| 74 | YBX1 | 5929 2389 | 18178 | -1.746 | -0.0401 | No | ||
| 75 | ZNF653 | 11459 | 18493 | -2.580 | 0.0066 | No |