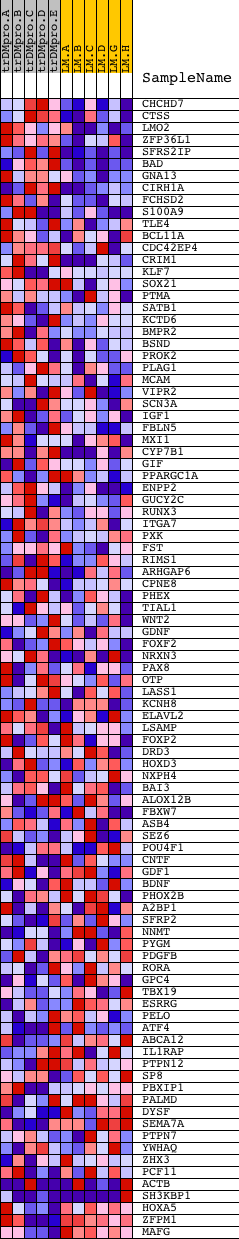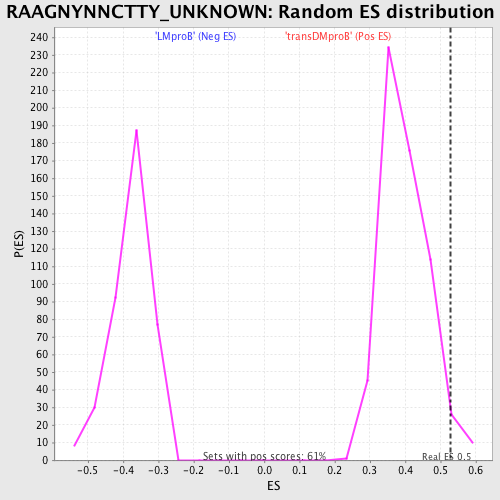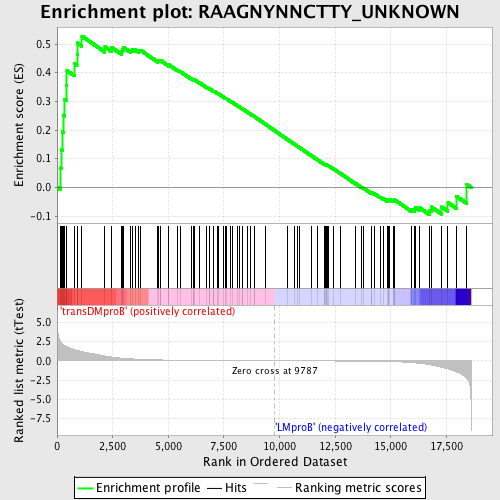
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | RAAGNYNNCTTY_UNKNOWN |
| Enrichment Score (ES) | 0.5275931 |
| Normalized Enrichment Score (NES) | 1.3236885 |
| Nominal p-value | 0.0330033 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CHCHD7 | 16280 | 135 | 2.545 | 0.0676 | Yes | ||
| 2 | CTSS | 15505 | 193 | 2.277 | 0.1315 | Yes | ||
| 3 | LMO2 | 9281 | 231 | 2.187 | 0.1938 | Yes | ||
| 4 | ZFP36L1 | 4453 4454 | 276 | 2.073 | 0.2524 | Yes | ||
| 5 | SFRS2IP | 7794 13009 | 348 | 1.949 | 0.3059 | Yes | ||
| 6 | BAD | 24000 | 423 | 1.823 | 0.3555 | Yes | ||
| 7 | GNA13 | 20617 | 442 | 1.793 | 0.4073 | Yes | ||
| 8 | CIRH1A | 18478 | 780 | 1.448 | 0.4317 | Yes | ||
| 9 | FCHSD2 | 18172 | 895 | 1.347 | 0.4652 | Yes | ||
| 10 | S100A9 | 9774 | 908 | 1.335 | 0.5038 | Yes | ||
| 11 | TLE4 | 23720 3697 | 1115 | 1.187 | 0.5276 | Yes | ||
| 12 | BCL11A | 4691 | 2149 | 0.616 | 0.4899 | No | ||
| 13 | CDC42EP4 | 20163 | 2441 | 0.492 | 0.4887 | No | ||
| 14 | CRIM1 | 403 | 2903 | 0.338 | 0.4738 | No | ||
| 15 | KLF7 | 8205 | 2922 | 0.332 | 0.4826 | No | ||
| 16 | SOX21 | 27 10278 2778 | 2995 | 0.311 | 0.4878 | No | ||
| 17 | PTMA | 9657 | 3303 | 0.245 | 0.4785 | No | ||
| 18 | SATB1 | 5408 | 3397 | 0.230 | 0.4802 | No | ||
| 19 | KCTD6 | 22098 | 3521 | 0.211 | 0.4798 | No | ||
| 20 | BMPR2 | 14241 4081 | 3678 | 0.190 | 0.4770 | No | ||
| 21 | BSND | 15821 | 3755 | 0.182 | 0.4782 | No | ||
| 22 | PROK2 | 17057 | 4524 | 0.110 | 0.4400 | No | ||
| 23 | PLAG1 | 7148 15949 12161 | 4540 | 0.109 | 0.4424 | No | ||
| 24 | MCAM | 3109 19479 | 4634 | 0.104 | 0.4404 | No | ||
| 25 | VIPR2 | 21125 | 4668 | 0.102 | 0.4416 | No | ||
| 26 | SCN3A | 14573 | 5013 | 0.085 | 0.4255 | No | ||
| 27 | IGF1 | 3352 9156 3409 | 5014 | 0.085 | 0.4280 | No | ||
| 28 | FBLN5 | 21002 | 5419 | 0.071 | 0.4083 | No | ||
| 29 | MXI1 | 23813 5139 | 5527 | 0.067 | 0.4045 | No | ||
| 30 | CYP7B1 | 15373 | 6038 | 0.053 | 0.3785 | No | ||
| 31 | GIF | 23923 | 6119 | 0.051 | 0.3757 | No | ||
| 32 | PPARGC1A | 16533 | 6137 | 0.051 | 0.3763 | No | ||
| 33 | ENPP2 | 9548 | 6196 | 0.049 | 0.3746 | No | ||
| 34 | GUCY2C | 1140 16954 | 6417 | 0.045 | 0.3640 | No | ||
| 35 | RUNX3 | 8702 | 6703 | 0.039 | 0.3498 | No | ||
| 36 | ITGA7 | 19835 | 6832 | 0.037 | 0.3440 | No | ||
| 37 | PXK | 26 | 6852 | 0.037 | 0.3441 | No | ||
| 38 | FST | 8985 | 7045 | 0.034 | 0.3347 | No | ||
| 39 | RIMS1 | 8575 | 7049 | 0.034 | 0.3355 | No | ||
| 40 | ARHGAP6 | 24207 | 7219 | 0.031 | 0.3273 | No | ||
| 41 | CPNE8 | 22156 | 7251 | 0.030 | 0.3265 | No | ||
| 42 | PHEX | 24024 | 7495 | 0.027 | 0.3142 | No | ||
| 43 | TIAL1 | 996 5753 | 7580 | 0.026 | 0.3104 | No | ||
| 44 | WNT2 | 17212 | 7603 | 0.025 | 0.3099 | No | ||
| 45 | GDNF | 22514 2275 | 7809 | 0.022 | 0.2995 | No | ||
| 46 | FOXF2 | 21677 | 7897 | 0.021 | 0.2955 | No | ||
| 47 | NRXN3 | 2153 5196 | 8107 | 0.018 | 0.2847 | No | ||
| 48 | PAX8 | 2791 14671 | 8197 | 0.017 | 0.2804 | No | ||
| 49 | OTP | 21582 | 8333 | 0.016 | 0.2736 | No | ||
| 50 | LASS1 | 3911 13566 | 8346 | 0.016 | 0.2734 | No | ||
| 51 | KCNH8 | 23197 | 8562 | 0.013 | 0.2622 | No | ||
| 52 | ELAVL2 | 15839 | 8685 | 0.012 | 0.2560 | No | ||
| 53 | LSAMP | 1690 6500 | 8861 | 0.010 | 0.2468 | No | ||
| 54 | FOXP2 | 4312 17523 8523 | 8877 | 0.010 | 0.2463 | No | ||
| 55 | DRD3 | 22750 | 9363 | 0.005 | 0.2202 | No | ||
| 56 | HOXD3 | 9115 | 10347 | -0.006 | 0.1674 | No | ||
| 57 | NXPH4 | 19602 | 10647 | -0.009 | 0.1515 | No | ||
| 58 | BAI3 | 5580 | 10797 | -0.011 | 0.1438 | No | ||
| 59 | ALOX12B | 20825 | 10896 | -0.012 | 0.1388 | No | ||
| 60 | FBXW7 | 1805 11928 | 11412 | -0.018 | 0.1115 | No | ||
| 61 | ASB4 | 990 17529 | 11682 | -0.021 | 0.0976 | No | ||
| 62 | SEZ6 | 20764 | 11999 | -0.025 | 0.0813 | No | ||
| 63 | POU4F1 | 21727 | 12034 | -0.026 | 0.0802 | No | ||
| 64 | CNTF | 3741 8761 3680 3736 | 12085 | -0.026 | 0.0783 | No | ||
| 65 | GDF1 | 4766 | 12095 | -0.026 | 0.0786 | No | ||
| 66 | BDNF | 14926 2797 | 12118 | -0.027 | 0.0782 | No | ||
| 67 | PHOX2B | 16524 | 12143 | -0.027 | 0.0777 | No | ||
| 68 | A2BP1 | 11212 1652 1689 | 12196 | -0.028 | 0.0757 | No | ||
| 69 | SFRP2 | 15564 | 12400 | -0.031 | 0.0656 | No | ||
| 70 | NNMT | 19131 | 12747 | -0.036 | 0.0480 | No | ||
| 71 | PYGM | 23792 | 13433 | -0.049 | 0.0125 | No | ||
| 72 | PDGFB | 22199 2311 | 13689 | -0.055 | 0.0004 | No | ||
| 73 | RORA | 3019 5388 | 13778 | -0.058 | -0.0027 | No | ||
| 74 | GPC4 | 24153 | 14110 | -0.067 | -0.0186 | No | ||
| 75 | TBX19 | 13780 | 14147 | -0.068 | -0.0185 | No | ||
| 76 | ESRRG | 6447 | 14257 | -0.072 | -0.0223 | No | ||
| 77 | PELO | 8360 | 14521 | -0.082 | -0.0341 | No | ||
| 78 | ATF4 | 22417 2239 | 14672 | -0.090 | -0.0395 | No | ||
| 79 | ABCA12 | 13931 | 14842 | -0.099 | -0.0457 | No | ||
| 80 | IL1RAP | 1653 1676 4917 1748 1712 9173 | 14876 | -0.101 | -0.0445 | No | ||
| 81 | PTPN12 | 16597 3502 3586 3665 | 14881 | -0.101 | -0.0418 | No | ||
| 82 | SP8 | 21122 | 14956 | -0.107 | -0.0426 | No | ||
| 83 | PBXIP1 | 6028 | 15099 | -0.119 | -0.0468 | No | ||
| 84 | PALMD | 15181 | 15150 | -0.122 | -0.0459 | No | ||
| 85 | DYSF | 6510 | 15167 | -0.124 | -0.0431 | No | ||
| 86 | SEMA7A | 19426 9803 | 15940 | -0.238 | -0.0778 | No | ||
| 87 | PTPN7 | 11500 | 16073 | -0.271 | -0.0770 | No | ||
| 88 | YWHAQ | 10369 | 16094 | -0.276 | -0.0699 | No | ||
| 89 | ZHX3 | 6800 | 16282 | -0.328 | -0.0704 | No | ||
| 90 | PCF11 | 121 | 16733 | -0.505 | -0.0798 | No | ||
| 91 | ACTB | 8534 337 337 338 | 16817 | -0.542 | -0.0683 | No | ||
| 92 | SH3KBP1 | 2631 12206 7189 | 17281 | -0.847 | -0.0684 | No | ||
| 93 | HOXA5 | 17149 | 17569 | -1.050 | -0.0530 | No | ||
| 94 | ZFPM1 | 18439 | 17952 | -1.445 | -0.0311 | No | ||
| 95 | MAFG | 5060 | 18420 | -2.276 | 0.0106 | No |