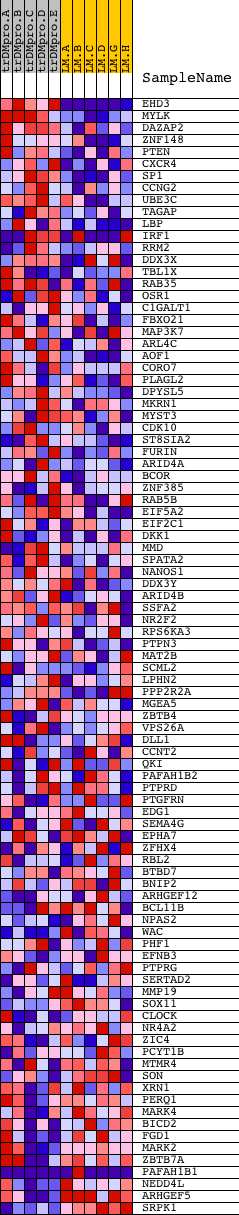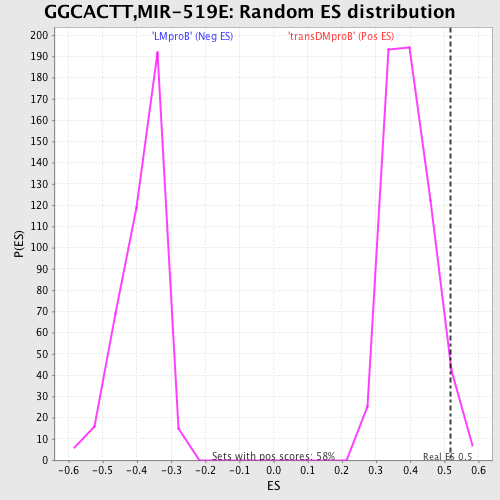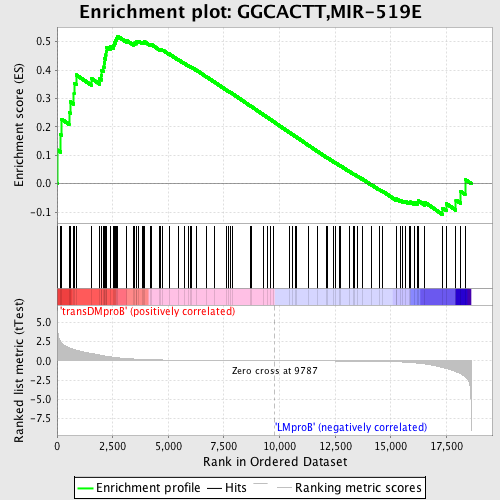
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | GGCACTT,MIR-519E |
| Enrichment Score (ES) | 0.5194837 |
| Normalized Enrichment Score (NES) | 1.3161727 |
| Nominal p-value | 0.025728988 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EHD3 | 23159 | 11 | 4.731 | 0.1186 | Yes | ||
| 2 | MYLK | 22778 4213 | 154 | 2.443 | 0.1724 | Yes | ||
| 3 | DAZAP2 | 10758 | 198 | 2.266 | 0.2272 | Yes | ||
| 4 | ZNF148 | 5949 | 540 | 1.670 | 0.2509 | Yes | ||
| 5 | PTEN | 5305 | 607 | 1.620 | 0.2881 | Yes | ||
| 6 | CXCR4 | 13844 | 758 | 1.469 | 0.3170 | Yes | ||
| 7 | SP1 | 9852 | 765 | 1.462 | 0.3535 | Yes | ||
| 8 | CCNG2 | 16790 | 863 | 1.377 | 0.3830 | Yes | ||
| 9 | UBE3C | 16896 | 1547 | 0.946 | 0.3699 | Yes | ||
| 10 | TAGAP | 6888 | 1913 | 0.740 | 0.3689 | Yes | ||
| 11 | LBP | 14760 | 1990 | 0.694 | 0.3823 | Yes | ||
| 12 | IRF1 | 1336 1258 1433 9182 | 2007 | 0.688 | 0.3987 | Yes | ||
| 13 | RRM2 | 5401 5400 | 2087 | 0.649 | 0.4108 | Yes | ||
| 14 | DDX3X | 24379 | 2114 | 0.640 | 0.4256 | Yes | ||
| 15 | TBL1X | 5628 | 2137 | 0.623 | 0.4401 | Yes | ||
| 16 | RAB35 | 13386 | 2174 | 0.598 | 0.4532 | Yes | ||
| 17 | OSR1 | 18971 | 2198 | 0.585 | 0.4667 | Yes | ||
| 18 | C1GALT1 | 8246 | 2225 | 0.572 | 0.4797 | Yes | ||
| 19 | FBXO21 | 16726 | 2387 | 0.507 | 0.4838 | Yes | ||
| 20 | MAP3K7 | 16255 | 2546 | 0.451 | 0.4866 | Yes | ||
| 21 | ARL4C | 13888 | 2562 | 0.444 | 0.4970 | Yes | ||
| 22 | AOF1 | 5746 3207 3284 | 2634 | 0.420 | 0.5037 | Yes | ||
| 23 | CORO7 | 22677 | 2660 | 0.409 | 0.5127 | Yes | ||
| 24 | PLAGL2 | 7055 | 2716 | 0.387 | 0.5195 | Yes | ||
| 25 | DPYSL5 | 92 | 3135 | 0.279 | 0.5039 | No | ||
| 26 | MKRN1 | 17178 | 3439 | 0.223 | 0.4932 | No | ||
| 27 | MYST3 | 18651 | 3498 | 0.214 | 0.4955 | No | ||
| 28 | CDK10 | 3818 6157 10635 3894 | 3569 | 0.203 | 0.4968 | No | ||
| 29 | ST8SIA2 | 17797 | 3580 | 0.201 | 0.5013 | No | ||
| 30 | FURIN | 9538 | 3671 | 0.191 | 0.5013 | No | ||
| 31 | ARID4A | 2080 6215 | 3819 | 0.173 | 0.4977 | No | ||
| 32 | BCOR | 12934 2561 | 3895 | 0.162 | 0.4977 | No | ||
| 33 | ZNF385 | 22106 2297 | 3921 | 0.159 | 0.5004 | No | ||
| 34 | RAB5B | 359 19591 3430 | 4184 | 0.134 | 0.4896 | No | ||
| 35 | EIF5A2 | 15622 | 4255 | 0.127 | 0.4891 | No | ||
| 36 | EIF2C1 | 10672 | 4619 | 0.105 | 0.4721 | No | ||
| 37 | DKK1 | 23700 | 4659 | 0.103 | 0.4726 | No | ||
| 38 | MMD | 20706 | 4744 | 0.097 | 0.4705 | No | ||
| 39 | SPATA2 | 6451 6450 | 5030 | 0.084 | 0.4572 | No | ||
| 40 | NANOS1 | 6858 | 5433 | 0.070 | 0.4373 | No | ||
| 41 | DDX3Y | 6507 4593 | 5727 | 0.061 | 0.4230 | No | ||
| 42 | ARID4B | 21705 3233 | 5891 | 0.057 | 0.4157 | No | ||
| 43 | SSFA2 | 7692 | 5989 | 0.054 | 0.4118 | No | ||
| 44 | NR2F2 | 8619 409 | 6037 | 0.053 | 0.4106 | No | ||
| 45 | RPS6KA3 | 8490 | 6252 | 0.048 | 0.4003 | No | ||
| 46 | PTPN3 | 9661 | 6728 | 0.039 | 0.3756 | No | ||
| 47 | MAT2B | 20494 1431 | 7067 | 0.033 | 0.3582 | No | ||
| 48 | SCML2 | 4221 | 7607 | 0.025 | 0.3297 | No | ||
| 49 | LPHN2 | 13619 | 7685 | 0.024 | 0.3262 | No | ||
| 50 | PPP2R2A | 3222 21774 | 7792 | 0.023 | 0.3210 | No | ||
| 51 | MGEA5 | 8001 | 7892 | 0.021 | 0.3162 | No | ||
| 52 | ZBTB4 | 13267 | 8682 | 0.012 | 0.2739 | No | ||
| 53 | VPS26A | 19752 3333 | 8750 | 0.011 | 0.2706 | No | ||
| 54 | DLL1 | 23119 | 9289 | 0.005 | 0.2417 | No | ||
| 55 | CCNT2 | 3993 13079 | 9444 | 0.004 | 0.2335 | No | ||
| 56 | QKI | 23128 5340 | 9609 | 0.002 | 0.2247 | No | ||
| 57 | PAFAH1B2 | 5221 9525 | 9714 | 0.001 | 0.2191 | No | ||
| 58 | PTPRD | 11554 5330 | 10467 | -0.007 | 0.1787 | No | ||
| 59 | PTGFRN | 15224 | 10566 | -0.008 | 0.1736 | No | ||
| 60 | EDG1 | 15187 | 10725 | -0.010 | 0.1653 | No | ||
| 61 | SEMA4G | 11182 | 10764 | -0.010 | 0.1635 | No | ||
| 62 | EPHA7 | 8909 4674 | 11300 | -0.016 | 0.1350 | No | ||
| 63 | ZFHX4 | 8158 | 11700 | -0.022 | 0.1140 | No | ||
| 64 | RBL2 | 18530 3781 | 12106 | -0.027 | 0.0929 | No | ||
| 65 | BTBD7 | 20998 | 12156 | -0.027 | 0.0909 | No | ||
| 66 | BNIP2 | 179 19388 | 12440 | -0.031 | 0.0764 | No | ||
| 67 | ARHGEF12 | 19156 | 12493 | -0.032 | 0.0744 | No | ||
| 68 | BCL11B | 7190 | 12709 | -0.035 | 0.0637 | No | ||
| 69 | NPAS2 | 5186 | 12759 | -0.036 | 0.0620 | No | ||
| 70 | WAC | 5905 5906 | 13134 | -0.043 | 0.0429 | No | ||
| 71 | PHF1 | 23324 1551 | 13321 | -0.047 | 0.0340 | No | ||
| 72 | EFNB3 | 20391 | 13374 | -0.048 | 0.0324 | No | ||
| 73 | PTPRG | 22097 3151 | 13521 | -0.051 | 0.0258 | No | ||
| 74 | SERTAD2 | 12200 7182 | 13711 | -0.056 | 0.0170 | No | ||
| 75 | MMP19 | 7191 | 14127 | -0.068 | -0.0037 | No | ||
| 76 | SOX11 | 5477 | 14475 | -0.081 | -0.0204 | No | ||
| 77 | CLOCK | 8749 16507 4530 | 14631 | -0.087 | -0.0266 | No | ||
| 78 | NR4A2 | 5203 | 15242 | -0.131 | -0.0562 | No | ||
| 79 | ZIC4 | 5992 | 15258 | -0.132 | -0.0537 | No | ||
| 80 | PCYT1B | 24289 | 15413 | -0.149 | -0.0582 | No | ||
| 81 | MTMR4 | 20712 | 15544 | -0.164 | -0.0611 | No | ||
| 82 | SON | 5473 1657 1684 | 15648 | -0.181 | -0.0621 | No | ||
| 83 | XRN1 | 10797 | 15825 | -0.214 | -0.0662 | No | ||
| 84 | PERQ1 | 7168 | 15898 | -0.227 | -0.0644 | No | ||
| 85 | MARK4 | 10572 | 16044 | -0.263 | -0.0656 | No | ||
| 86 | BICD2 | 8048 | 16185 | -0.301 | -0.0656 | No | ||
| 87 | FGD1 | 24228 | 16229 | -0.312 | -0.0601 | No | ||
| 88 | MARK2 | 8899 | 16521 | -0.412 | -0.0654 | No | ||
| 89 | ZBTB7A | 5004 | 17323 | -0.885 | -0.0863 | No | ||
| 90 | PAFAH1B1 | 1340 5220 9524 | 17497 | -1.002 | -0.0704 | No | ||
| 91 | NEDD4L | 8192 | 17926 | -1.415 | -0.0579 | No | ||
| 92 | ARHGEF5 | 12025 | 18132 | -1.668 | -0.0269 | No | ||
| 93 | SRPK1 | 23049 | 18339 | -2.104 | 0.0150 | No |