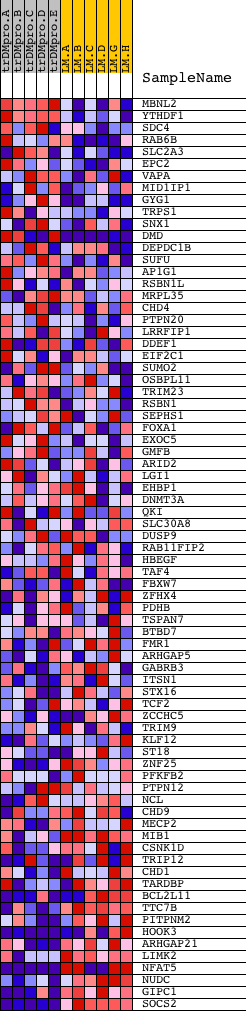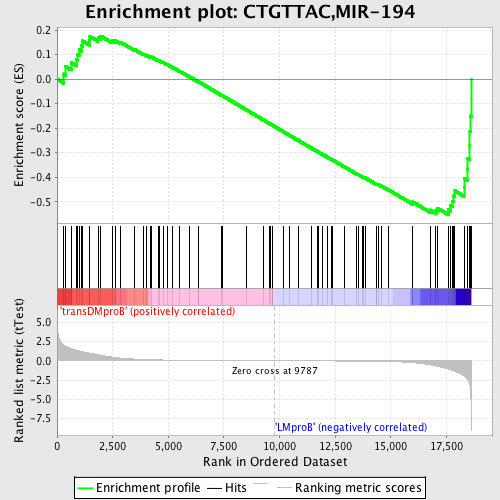
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | CTGTTAC,MIR-194 |
| Enrichment Score (ES) | -0.5518596 |
| Normalized Enrichment Score (NES) | -1.4266552 |
| Nominal p-value | 0.023980815 |
| FDR q-value | 0.66080177 |
| FWER p-Value | 0.976 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MBNL2 | 21932 | 305 | 2.027 | 0.0208 | No | ||
| 2 | YTHDF1 | 14311 | 390 | 1.869 | 0.0506 | No | ||
| 3 | SDC4 | 14349 | 632 | 1.600 | 0.0670 | No | ||
| 4 | RAB6B | 11289 | 879 | 1.361 | 0.0787 | No | ||
| 5 | SLC2A3 | 17010 | 933 | 1.314 | 0.0999 | No | ||
| 6 | EPC2 | 15015 | 992 | 1.273 | 0.1202 | No | ||
| 7 | VAPA | 22908 | 1105 | 1.193 | 0.1361 | No | ||
| 8 | MID1IP1 | 24381 | 1145 | 1.172 | 0.1555 | No | ||
| 9 | GYG1 | 1820 15371 | 1458 | 0.988 | 0.1568 | No | ||
| 10 | TRPS1 | 8195 | 1475 | 0.982 | 0.1740 | No | ||
| 11 | SNX1 | 19076 | 1837 | 0.783 | 0.1689 | No | ||
| 12 | DMD | 24295 2647 | 1963 | 0.710 | 0.1752 | No | ||
| 13 | DEPDC1B | 5757 | 2476 | 0.480 | 0.1564 | No | ||
| 14 | SUFU | 6283 | 2627 | 0.423 | 0.1560 | No | ||
| 15 | AP1G1 | 4392 | 2860 | 0.348 | 0.1499 | No | ||
| 16 | RSBN1L | 16598 | 3494 | 0.215 | 0.1197 | No | ||
| 17 | MRPL35 | 7267 | 3885 | 0.164 | 0.1017 | No | ||
| 18 | CHD4 | 8418 17281 4225 | 4036 | 0.148 | 0.0964 | No | ||
| 19 | PTPN20 | 22049 | 4201 | 0.132 | 0.0899 | No | ||
| 20 | LRRFIP1 | 14191 4007 3965 | 4236 | 0.129 | 0.0905 | No | ||
| 21 | DDEF1 | 22282 2253 | 4538 | 0.109 | 0.0762 | No | ||
| 22 | EIF2C1 | 10672 | 4619 | 0.105 | 0.0739 | No | ||
| 23 | SUMO2 | 1293 9319 | 4762 | 0.096 | 0.0680 | No | ||
| 24 | OSBPL11 | 22781 | 4939 | 0.088 | 0.0601 | No | ||
| 25 | TRIM23 | 8165 3230 | 5205 | 0.078 | 0.0472 | No | ||
| 26 | RSBN1 | 6035 | 5501 | 0.068 | 0.0325 | No | ||
| 27 | SEPHS1 | 15125 | 5940 | 0.056 | 0.0099 | No | ||
| 28 | FOXA1 | 21060 | 6363 | 0.046 | -0.0120 | No | ||
| 29 | EXOC5 | 8370 | 6374 | 0.045 | -0.0117 | No | ||
| 30 | GMFB | 21861 | 7408 | 0.028 | -0.0669 | No | ||
| 31 | ARID2 | 8062 13378 | 7433 | 0.028 | -0.0677 | No | ||
| 32 | LGI1 | 23867 | 8494 | 0.014 | -0.1246 | No | ||
| 33 | EHBP1 | 20516 | 9280 | 0.005 | -0.1668 | No | ||
| 34 | DNMT3A | 2167 21330 | 9546 | 0.003 | -0.1811 | No | ||
| 35 | QKI | 23128 5340 | 9609 | 0.002 | -0.1844 | No | ||
| 36 | SLC30A8 | 10737 22478 | 9690 | 0.001 | -0.1887 | No | ||
| 37 | DUSP9 | 24307 | 9702 | 0.001 | -0.1892 | No | ||
| 38 | RAB11FIP2 | 23635 | 10160 | -0.004 | -0.2138 | No | ||
| 39 | HBEGF | 23457 | 10446 | -0.007 | -0.2290 | No | ||
| 40 | TAF4 | 14319 | 10838 | -0.011 | -0.2499 | No | ||
| 41 | FBXW7 | 1805 11928 | 11412 | -0.018 | -0.2805 | No | ||
| 42 | ZFHX4 | 8158 | 11700 | -0.022 | -0.2956 | No | ||
| 43 | PDHB | 12670 7548 | 11762 | -0.022 | -0.2985 | No | ||
| 44 | TSPAN7 | 24382 | 11919 | -0.024 | -0.3064 | No | ||
| 45 | BTBD7 | 20998 | 12156 | -0.027 | -0.3187 | No | ||
| 46 | FMR1 | 24321 | 12334 | -0.030 | -0.3277 | No | ||
| 47 | ARHGAP5 | 4412 8625 | 12391 | -0.030 | -0.3301 | No | ||
| 48 | GABRB3 | 4747 | 12896 | -0.039 | -0.3566 | No | ||
| 49 | ITSN1 | 9196 | 13462 | -0.050 | -0.3861 | No | ||
| 50 | STX16 | 10464 | 13532 | -0.052 | -0.3889 | No | ||
| 51 | TCF2 | 1395 20727 | 13715 | -0.056 | -0.3977 | No | ||
| 52 | ZCCHC5 | 24081 | 13781 | -0.058 | -0.4001 | No | ||
| 53 | TRIM9 | 2118 8076 13577 8234 | 13851 | -0.060 | -0.4028 | No | ||
| 54 | KLF12 | 4960 2637 21732 9228 | 14352 | -0.076 | -0.4283 | No | ||
| 55 | ST18 | 14298 | 14369 | -0.077 | -0.4278 | No | ||
| 56 | ZNF25 | 17025 | 14430 | -0.079 | -0.4296 | No | ||
| 57 | PFKFB2 | 4116 5242 | 14574 | -0.084 | -0.4357 | No | ||
| 58 | PTPN12 | 16597 3502 3586 3665 | 14881 | -0.101 | -0.4504 | No | ||
| 59 | NCL | 5153 13899 | 15981 | -0.247 | -0.5051 | No | ||
| 60 | CHD9 | 6784 11553 18531 | 15987 | -0.249 | -0.5008 | No | ||
| 61 | MECP2 | 5088 | 16772 | -0.524 | -0.5335 | No | ||
| 62 | MIB1 | 5908 | 17009 | -0.652 | -0.5342 | Yes | ||
| 63 | CSNK1D | 4181 8352 1387 | 17102 | -0.712 | -0.5261 | Yes | ||
| 64 | TRIP12 | 4813 9055 9054 | 17580 | -1.059 | -0.5324 | Yes | ||
| 65 | CHD1 | 23377 | 17684 | -1.173 | -0.5164 | Yes | ||
| 66 | TARDBP | 10520 6066 | 17762 | -1.243 | -0.4978 | Yes | ||
| 67 | BCL2L11 | 2790 14861 | 17805 | -1.285 | -0.4764 | Yes | ||
| 68 | TTC7B | 21006 | 17882 | -1.357 | -0.4556 | Yes | ||
| 69 | PITPNM2 | 16371 | 18291 | -1.975 | -0.4414 | Yes | ||
| 70 | HOOK3 | 18903 6734 | 18298 | -1.994 | -0.4051 | Yes | ||
| 71 | ARHGAP21 | 12929 | 18453 | -2.431 | -0.3687 | Yes | ||
| 72 | LIMK2 | 9278 1296 | 18462 | -2.463 | -0.3239 | Yes | ||
| 73 | NFAT5 | 3921 7037 12036 | 18552 | -3.129 | -0.2713 | Yes | ||
| 74 | NUDC | 9495 | 18556 | -3.153 | -0.2136 | Yes | ||
| 75 | GIPC1 | 3841 3882 18552 | 18580 | -3.506 | -0.1504 | Yes | ||
| 76 | SOCS2 | 5694 | 18614 | -8.294 | 0.0001 | Yes |