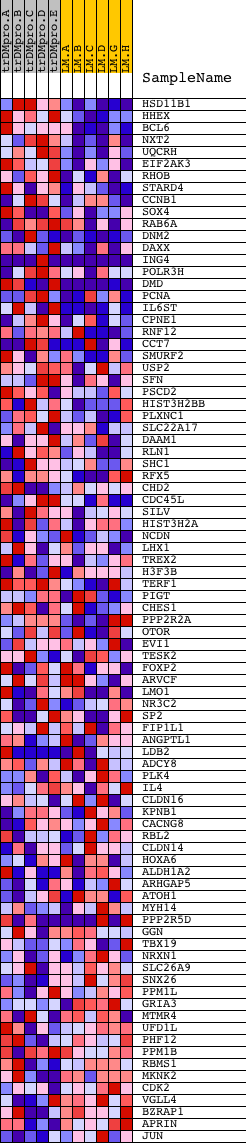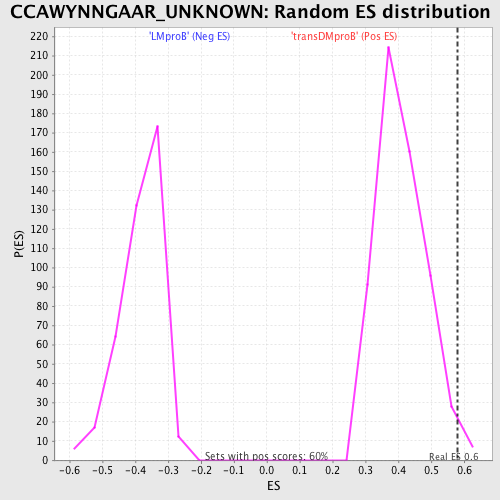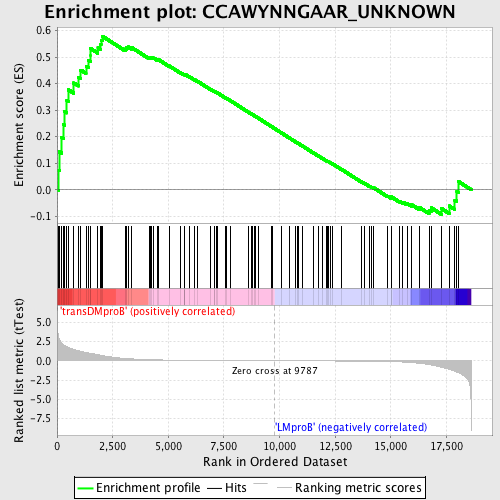
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | CCAWYNNGAAR_UNKNOWN |
| Enrichment Score (ES) | 0.5789006 |
| Normalized Enrichment Score (NES) | 1.4098183 |
| Nominal p-value | 0.015100671 |
| FDR q-value | 0.81565666 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HSD11B1 | 9125 4019 | 67 | 2.980 | 0.0741 | Yes | ||
| 2 | HHEX | 23872 | 102 | 2.715 | 0.1430 | Yes | ||
| 3 | BCL6 | 22624 | 205 | 2.250 | 0.1962 | Yes | ||
| 4 | NXT2 | 10685 | 290 | 2.053 | 0.2452 | Yes | ||
| 5 | UQCRH | 12378 | 330 | 1.989 | 0.2949 | Yes | ||
| 6 | EIF2AK3 | 17421 | 437 | 1.796 | 0.3360 | Yes | ||
| 7 | RHOB | 4411 | 510 | 1.704 | 0.3766 | Yes | ||
| 8 | STARD4 | 5011 | 747 | 1.482 | 0.4025 | Yes | ||
| 9 | CCNB1 | 11201 21362 | 963 | 1.293 | 0.4246 | Yes | ||
| 10 | SOX4 | 5479 | 1064 | 1.224 | 0.4511 | Yes | ||
| 11 | RAB6A | 18173 3905 | 1300 | 1.066 | 0.4662 | Yes | ||
| 12 | DNM2 | 4635 3103 | 1404 | 1.002 | 0.4868 | Yes | ||
| 13 | DAXX | 23290 | 1495 | 0.973 | 0.5073 | Yes | ||
| 14 | ING4 | 6602 1155 1102 11371 | 1499 | 0.972 | 0.5325 | Yes | ||
| 15 | POLR3H | 13460 8135 | 1836 | 0.783 | 0.5347 | Yes | ||
| 16 | DMD | 24295 2647 | 1963 | 0.710 | 0.5464 | Yes | ||
| 17 | PCNA | 9535 | 1977 | 0.701 | 0.5640 | Yes | ||
| 18 | IL6ST | 4920 | 2029 | 0.676 | 0.5789 | Yes | ||
| 19 | CPNE1 | 11186 | 3068 | 0.295 | 0.5306 | No | ||
| 20 | RNF12 | 5384 | 3108 | 0.284 | 0.5359 | No | ||
| 21 | CCT7 | 17385 | 3193 | 0.267 | 0.5383 | No | ||
| 22 | SMURF2 | 185 | 3355 | 0.237 | 0.5358 | No | ||
| 23 | USP2 | 19480 3052 3043 | 4152 | 0.137 | 0.4964 | No | ||
| 24 | SFN | 7060 12062 | 4206 | 0.132 | 0.4970 | No | ||
| 25 | PSCD2 | 17826 | 4245 | 0.128 | 0.4983 | No | ||
| 26 | HIST3H2BB | 11796 | 4334 | 0.121 | 0.4967 | No | ||
| 27 | PLXNC1 | 7056 | 4516 | 0.110 | 0.4898 | No | ||
| 28 | SLC22A17 | 21830 3096 | 4546 | 0.109 | 0.4910 | No | ||
| 29 | DAAM1 | 5536 | 5040 | 0.084 | 0.4666 | No | ||
| 30 | RLN1 | 9726 | 5547 | 0.066 | 0.4410 | No | ||
| 31 | SHC1 | 9813 9812 5430 | 5712 | 0.062 | 0.4338 | No | ||
| 32 | RFX5 | 7018 | 5714 | 0.062 | 0.4353 | No | ||
| 33 | CHD2 | 10847 | 5731 | 0.061 | 0.4361 | No | ||
| 34 | CDC45L | 22642 1752 | 5968 | 0.055 | 0.4248 | No | ||
| 35 | SILV | 16130 | 6187 | 0.050 | 0.4143 | No | ||
| 36 | HIST3H2A | 6647 | 6325 | 0.046 | 0.4081 | No | ||
| 37 | NCDN | 15756 2487 6471 | 6890 | 0.036 | 0.3786 | No | ||
| 38 | LHX1 | 4995 | 7059 | 0.033 | 0.3704 | No | ||
| 39 | TREX2 | 24137 | 7071 | 0.033 | 0.3707 | No | ||
| 40 | H3F3B | 4839 | 7177 | 0.032 | 0.3659 | No | ||
| 41 | TERF1 | 14293 | 7215 | 0.031 | 0.3647 | No | ||
| 42 | PIGT | 8134 1 | 7549 | 0.026 | 0.3474 | No | ||
| 43 | CHES1 | 21007 11200 | 7601 | 0.025 | 0.3453 | No | ||
| 44 | PPP2R2A | 3222 21774 | 7792 | 0.023 | 0.3356 | No | ||
| 45 | OTOR | 14825 | 8617 | 0.013 | 0.2915 | No | ||
| 46 | EVI1 | 4689 8926 | 8754 | 0.011 | 0.2844 | No | ||
| 47 | TESK2 | 10504 | 8793 | 0.011 | 0.2827 | No | ||
| 48 | FOXP2 | 4312 17523 8523 | 8877 | 0.010 | 0.2784 | No | ||
| 49 | ARVCF | 8629 | 8917 | 0.010 | 0.2766 | No | ||
| 50 | LMO1 | 17685 | 9046 | 0.008 | 0.2699 | No | ||
| 51 | NR3C2 | 18562 13798 | 9628 | 0.002 | 0.2386 | No | ||
| 52 | SP2 | 8130 | 9663 | 0.001 | 0.2368 | No | ||
| 53 | FIP1L1 | 888 16826 | 10091 | -0.003 | 0.2138 | No | ||
| 54 | ANGPTL1 | 10195 8589 13064 | 10440 | -0.007 | 0.1952 | No | ||
| 55 | LDB2 | 16538 4989 | 10710 | -0.010 | 0.1810 | No | ||
| 56 | ADCY8 | 22281 | 10808 | -0.011 | 0.1760 | No | ||
| 57 | PLK4 | 1870 1874 | 10857 | -0.011 | 0.1737 | No | ||
| 58 | IL4 | 9174 | 11022 | -0.013 | 0.1652 | No | ||
| 59 | CLDN16 | 22802 | 11535 | -0.019 | 0.1381 | No | ||
| 60 | KPNB1 | 20274 | 11745 | -0.022 | 0.1274 | No | ||
| 61 | CACNG8 | 13498 | 11937 | -0.024 | 0.1177 | No | ||
| 62 | RBL2 | 18530 3781 | 12106 | -0.027 | 0.1094 | No | ||
| 63 | CLDN14 | 12081 | 12166 | -0.027 | 0.1069 | No | ||
| 64 | HOXA6 | 17150 | 12179 | -0.028 | 0.1070 | No | ||
| 65 | ALDH1A2 | 19386 | 12289 | -0.029 | 0.1018 | No | ||
| 66 | ARHGAP5 | 4412 8625 | 12391 | -0.030 | 0.0972 | No | ||
| 67 | ATOH1 | 4419 | 12796 | -0.037 | 0.0763 | No | ||
| 68 | MYH14 | 17845 | 13682 | -0.055 | 0.0300 | No | ||
| 69 | PPP2R5D | 962 1523 22957 10139 | 13803 | -0.059 | 0.0251 | No | ||
| 70 | GGN | 2472 1171 18312 | 14052 | -0.066 | 0.0134 | No | ||
| 71 | TBX19 | 13780 | 14147 | -0.068 | 0.0101 | No | ||
| 72 | NRXN1 | 1581 1575 22875 | 14220 | -0.071 | 0.0081 | No | ||
| 73 | SLC26A9 | 11560 980 4067 | 14868 | -0.101 | -0.0242 | No | ||
| 74 | SNX26 | 10582 | 15008 | -0.112 | -0.0288 | No | ||
| 75 | PPM1L | 10811 6316 | 15009 | -0.112 | -0.0259 | No | ||
| 76 | GRIA3 | 24349 11996 2649 | 15389 | -0.146 | -0.0426 | No | ||
| 77 | MTMR4 | 20712 | 15544 | -0.164 | -0.0466 | No | ||
| 78 | UFD1L | 22825 | 15737 | -0.198 | -0.0518 | No | ||
| 79 | PHF12 | 11192 6483 | 15922 | -0.234 | -0.0556 | No | ||
| 80 | PPM1B | 5280 1567 5281 | 16297 | -0.332 | -0.0671 | No | ||
| 81 | RBMS1 | 14580 | 16721 | -0.499 | -0.0770 | No | ||
| 82 | MKNK2 | 3299 9392 | 16815 | -0.542 | -0.0679 | No | ||
| 83 | CDK2 | 3438 3373 19592 3322 | 17283 | -0.848 | -0.0710 | No | ||
| 84 | VGLL4 | 10558 | 17625 | -1.096 | -0.0608 | No | ||
| 85 | BZRAP1 | 9882 | 17878 | -1.354 | -0.0391 | No | ||
| 86 | APRIN | 4145 | 17968 | -1.465 | -0.0057 | No | ||
| 87 | JUN | 15832 | 18032 | -1.559 | 0.0315 | No |