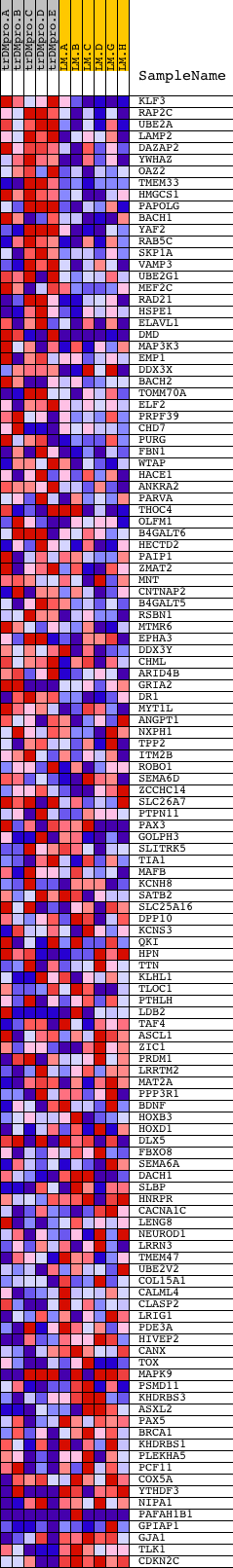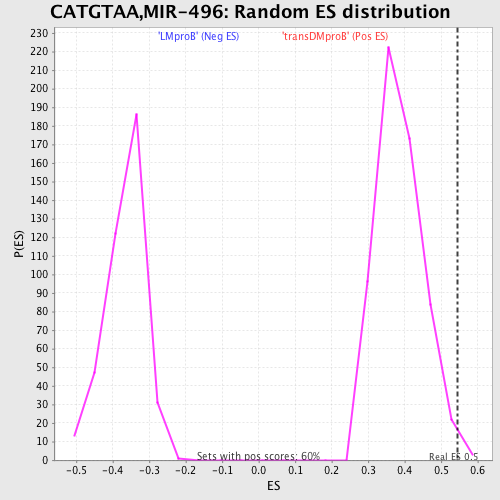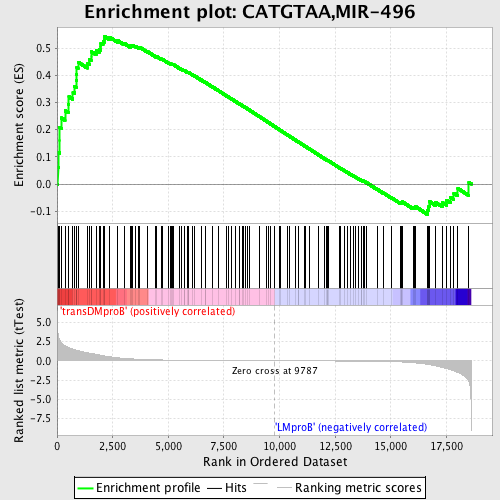
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | CATGTAA,MIR-496 |
| Enrichment Score (ES) | 0.54366845 |
| Normalized Enrichment Score (NES) | 1.4071096 |
| Nominal p-value | 0.006666667 |
| FDR q-value | 0.7351514 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | KLF3 | 4961 | 39 | 3.509 | 0.0607 | Yes | ||
| 2 | RAP2C | 24156 | 55 | 3.172 | 0.1166 | Yes | ||
| 3 | UBE2A | 24358 | 116 | 2.617 | 0.1602 | Yes | ||
| 4 | LAMP2 | 9267 2653 | 118 | 2.612 | 0.2069 | Yes | ||
| 5 | DAZAP2 | 10758 | 198 | 2.266 | 0.2432 | Yes | ||
| 6 | YWHAZ | 10370 | 356 | 1.942 | 0.2694 | Yes | ||
| 7 | OAZ2 | 9499 | 500 | 1.713 | 0.2923 | Yes | ||
| 8 | TMEM33 | 7497 3627 | 532 | 1.675 | 0.3206 | Yes | ||
| 9 | HMGCS1 | 9917 | 713 | 1.516 | 0.3380 | Yes | ||
| 10 | PAPOLG | 20512 | 786 | 1.445 | 0.3600 | Yes | ||
| 11 | BACH1 | 8647 | 860 | 1.379 | 0.3807 | Yes | ||
| 12 | YAF2 | 22155 | 877 | 1.362 | 0.4042 | Yes | ||
| 13 | RAB5C | 20226 | 887 | 1.356 | 0.4280 | Yes | ||
| 14 | SKP1A | 20889 | 943 | 1.308 | 0.4484 | Yes | ||
| 15 | VAMP3 | 15660 | 1351 | 1.026 | 0.4448 | Yes | ||
| 16 | UBE2G1 | 12472 7390 1353 | 1455 | 0.989 | 0.4569 | Yes | ||
| 17 | MEF2C | 3204 9378 | 1531 | 0.955 | 0.4699 | Yes | ||
| 18 | RAD21 | 22298 | 1532 | 0.955 | 0.4870 | Yes | ||
| 19 | HSPE1 | 9133 | 1756 | 0.828 | 0.4898 | Yes | ||
| 20 | ELAVL1 | 4888 | 1889 | 0.751 | 0.4961 | Yes | ||
| 21 | DMD | 24295 2647 | 1963 | 0.710 | 0.5048 | Yes | ||
| 22 | MAP3K3 | 20626 | 1967 | 0.706 | 0.5173 | Yes | ||
| 23 | EMP1 | 17260 | 2065 | 0.659 | 0.5239 | Yes | ||
| 24 | DDX3X | 24379 | 2114 | 0.640 | 0.5327 | Yes | ||
| 25 | BACH2 | 8648 | 2122 | 0.633 | 0.5437 | Yes | ||
| 26 | TOMM70A | 6608 | 2368 | 0.515 | 0.5396 | No | ||
| 27 | ELF2 | 1795 1856 12785 1882 | 2715 | 0.388 | 0.5279 | No | ||
| 28 | PRPF39 | 21259 | 3011 | 0.307 | 0.5174 | No | ||
| 29 | CHD7 | 16278 | 3311 | 0.244 | 0.5056 | No | ||
| 30 | PURG | 7943 | 3324 | 0.242 | 0.5093 | No | ||
| 31 | FBN1 | 14449 | 3403 | 0.229 | 0.5092 | No | ||
| 32 | WTAP | 7215 | 3510 | 0.212 | 0.5072 | No | ||
| 33 | HACE1 | 20034 | 3659 | 0.192 | 0.5027 | No | ||
| 34 | ANKRA2 | 21578 | 3707 | 0.186 | 0.5035 | No | ||
| 35 | PARVA | 18122 | 4082 | 0.144 | 0.4858 | No | ||
| 36 | THOC4 | 5710 10114 | 4437 | 0.115 | 0.4687 | No | ||
| 37 | OLFM1 | 2918 2971 | 4478 | 0.113 | 0.4686 | No | ||
| 38 | B4GALT6 | 7109 12115 | 4674 | 0.102 | 0.4598 | No | ||
| 39 | HECTD2 | 23876 3721 | 4733 | 0.099 | 0.4585 | No | ||
| 40 | PAIP1 | 21556 | 5012 | 0.085 | 0.4450 | No | ||
| 41 | ZMAT2 | 23591 | 5099 | 0.081 | 0.4418 | No | ||
| 42 | MNT | 20784 | 5133 | 0.080 | 0.4414 | No | ||
| 43 | CNTNAP2 | 17463 12406 | 5195 | 0.078 | 0.4395 | No | ||
| 44 | B4GALT5 | 14333 | 5215 | 0.077 | 0.4399 | No | ||
| 45 | RSBN1 | 6035 | 5501 | 0.068 | 0.4257 | No | ||
| 46 | MTMR6 | 21990 | 5610 | 0.064 | 0.4210 | No | ||
| 47 | EPHA3 | 22563 1643 | 5707 | 0.062 | 0.4169 | No | ||
| 48 | DDX3Y | 6507 4593 | 5727 | 0.061 | 0.4170 | No | ||
| 49 | CHML | 13741 | 5848 | 0.058 | 0.4115 | No | ||
| 50 | ARID4B | 21705 3233 | 5891 | 0.057 | 0.4103 | No | ||
| 51 | GRIA2 | 4802 | 5895 | 0.057 | 0.4111 | No | ||
| 52 | DR1 | 16762 | 6095 | 0.052 | 0.4013 | No | ||
| 53 | MYT1L | 5143 9441 | 6192 | 0.050 | 0.3970 | No | ||
| 54 | ANGPT1 | 8588 2260 22303 | 6469 | 0.044 | 0.3828 | No | ||
| 55 | NXPH1 | 17524 | 6679 | 0.040 | 0.3722 | No | ||
| 56 | TPP2 | 14257 | 6688 | 0.040 | 0.3725 | No | ||
| 57 | ITM2B | 9192 4935 21753 | 7004 | 0.034 | 0.3561 | No | ||
| 58 | ROBO1 | 9731 | 7231 | 0.031 | 0.3444 | No | ||
| 59 | SEMA6D | 2800 14876 90 | 7628 | 0.025 | 0.3234 | No | ||
| 60 | ZCCHC14 | 4731 | 7708 | 0.024 | 0.3196 | No | ||
| 61 | SLC26A7 | 5538 2372 | 7828 | 0.022 | 0.3136 | No | ||
| 62 | PTPN11 | 5326 16391 9660 | 8010 | 0.020 | 0.3041 | No | ||
| 63 | PAX3 | 13910 | 8195 | 0.017 | 0.2945 | No | ||
| 64 | GOLPH3 | 12388 7315 | 8219 | 0.017 | 0.2935 | No | ||
| 65 | SLITRK5 | 21939 | 8328 | 0.016 | 0.2880 | No | ||
| 66 | TIA1 | 5752 | 8394 | 0.015 | 0.2847 | No | ||
| 67 | MAFB | 14367 | 8450 | 0.014 | 0.2820 | No | ||
| 68 | KCNH8 | 23197 | 8562 | 0.013 | 0.2763 | No | ||
| 69 | SATB2 | 5604 | 8666 | 0.012 | 0.2709 | No | ||
| 70 | SLC25A16 | 20003 | 9082 | 0.008 | 0.2486 | No | ||
| 71 | DPP10 | 13850 | 9424 | 0.004 | 0.2302 | No | ||
| 72 | KCNS3 | 21109 | 9522 | 0.003 | 0.2250 | No | ||
| 73 | QKI | 23128 5340 | 9609 | 0.002 | 0.2204 | No | ||
| 74 | HPN | 17873 4087 | 9768 | 0.000 | 0.2119 | No | ||
| 75 | TTN | 14552 2743 14553 | 9976 | -0.002 | 0.2007 | No | ||
| 76 | KLHL1 | 21735 | 10023 | -0.003 | 0.1983 | No | ||
| 77 | TLOC1 | 12787 | 10368 | -0.006 | 0.1798 | No | ||
| 78 | PTHLH | 16931 | 10460 | -0.007 | 0.1750 | No | ||
| 79 | LDB2 | 16538 4989 | 10710 | -0.010 | 0.1617 | No | ||
| 80 | TAF4 | 14319 | 10838 | -0.011 | 0.1551 | No | ||
| 81 | ASCL1 | 19655 | 10848 | -0.011 | 0.1548 | No | ||
| 82 | ZIC1 | 19040 | 11139 | -0.015 | 0.1394 | No | ||
| 83 | PRDM1 | 19775 3337 | 11162 | -0.015 | 0.1384 | No | ||
| 84 | LRRTM2 | 4203 | 11358 | -0.017 | 0.1282 | No | ||
| 85 | MAT2A | 10553 10554 6099 | 11769 | -0.022 | 0.1064 | No | ||
| 86 | PPP3R1 | 9611 489 | 12037 | -0.026 | 0.0924 | No | ||
| 87 | BDNF | 14926 2797 | 12118 | -0.027 | 0.0886 | No | ||
| 88 | HOXB3 | 20685 | 12141 | -0.027 | 0.0879 | No | ||
| 89 | HOXD1 | 14981 | 12172 | -0.027 | 0.0868 | No | ||
| 90 | DLX5 | 17219 1058 | 12199 | -0.028 | 0.0859 | No | ||
| 91 | FBXO8 | 18616 3896 | 12700 | -0.035 | 0.0594 | No | ||
| 92 | SEMA6A | 23439 9802 | 12756 | -0.036 | 0.0571 | No | ||
| 93 | DACH1 | 4595 3371 | 12899 | -0.039 | 0.0501 | No | ||
| 94 | SLBP | 9822 | 13033 | -0.042 | 0.0437 | No | ||
| 95 | HNRPR | 13196 7909 2500 | 13199 | -0.044 | 0.0356 | No | ||
| 96 | CACNA1C | 1158 1166 24464 4463 8674 | 13332 | -0.047 | 0.0293 | No | ||
| 97 | LENG8 | 6114 | 13432 | -0.049 | 0.0248 | No | ||
| 98 | NEUROD1 | 14550 | 13552 | -0.052 | 0.0193 | No | ||
| 99 | LRRN3 | 21077 | 13663 | -0.055 | 0.0143 | No | ||
| 100 | TMEM47 | 5315 | 13692 | -0.056 | 0.0138 | No | ||
| 101 | UBE2V2 | 12888 | 13750 | -0.057 | 0.0117 | No | ||
| 102 | COL15A1 | 8764 | 13804 | -0.059 | 0.0099 | No | ||
| 103 | CALML4 | 19414 | 13920 | -0.062 | 0.0048 | No | ||
| 104 | CLASP2 | 13338 | 14387 | -0.077 | -0.0190 | No | ||
| 105 | LRIG1 | 9179 | 14667 | -0.090 | -0.0325 | No | ||
| 106 | PDE3A | 7045 | 15024 | -0.112 | -0.0498 | No | ||
| 107 | HIVEP2 | 9091 | 15437 | -0.151 | -0.0693 | No | ||
| 108 | CANX | 4474 | 15468 | -0.155 | -0.0682 | No | ||
| 109 | TOX | 15943 | 15481 | -0.156 | -0.0660 | No | ||
| 110 | MAPK9 | 1233 20903 1383 | 15505 | -0.159 | -0.0645 | No | ||
| 111 | PSMD11 | 12772 7600 | 15998 | -0.252 | -0.0866 | No | ||
| 112 | KHDRBS3 | 22462 | 16083 | -0.273 | -0.0862 | No | ||
| 113 | ASXL2 | 7957 | 16110 | -0.280 | -0.0826 | No | ||
| 114 | PAX5 | 15892 | 16649 | -0.461 | -0.1035 | No | ||
| 115 | BRCA1 | 20213 | 16654 | -0.466 | -0.0953 | No | ||
| 116 | KHDRBS1 | 5405 9778 | 16689 | -0.478 | -0.0886 | No | ||
| 117 | PLEKHA5 | 8453 | 16708 | -0.491 | -0.0808 | No | ||
| 118 | PCF11 | 121 | 16733 | -0.505 | -0.0731 | No | ||
| 119 | COX5A | 19431 | 16734 | -0.506 | -0.0640 | No | ||
| 120 | YTHDF3 | 15630 10469 6019 | 17016 | -0.654 | -0.0675 | No | ||
| 121 | NIPA1 | 6124 | 17320 | -0.884 | -0.0681 | No | ||
| 122 | PAFAH1B1 | 1340 5220 9524 | 17497 | -1.002 | -0.0597 | No | ||
| 123 | GPIAP1 | 2819 7014 | 17679 | -1.166 | -0.0486 | No | ||
| 124 | GJA1 | 20023 | 17834 | -1.315 | -0.0334 | No | ||
| 125 | TLK1 | 14563 | 18014 | -1.532 | -0.0157 | No | ||
| 126 | CDKN2C | 15806 2321 | 18509 | -2.692 | 0.0058 | No |