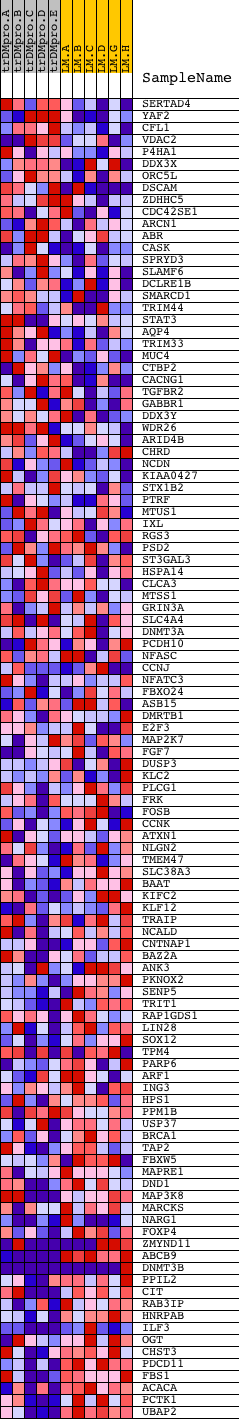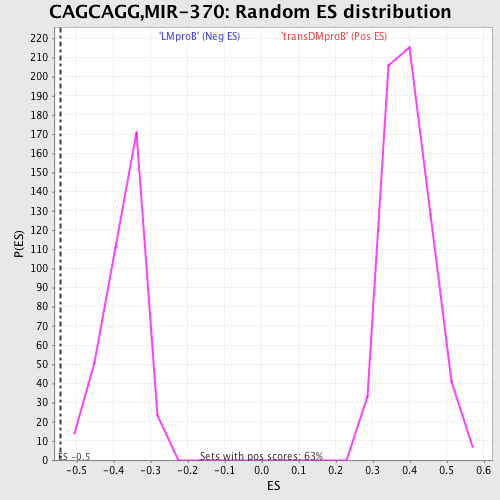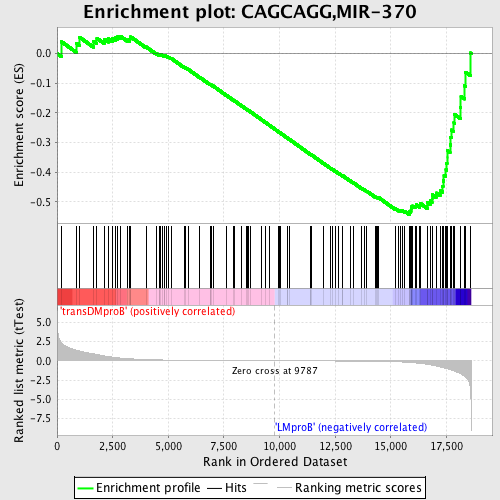
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | CAGCAGG,MIR-370 |
| Enrichment Score (ES) | -0.5429053 |
| Normalized Enrichment Score (NES) | -1.4578164 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.56283 |
| FWER p-Value | 0.939 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SERTAD4 | 13715 | 202 | 2.260 | 0.0400 | No | ||
| 2 | YAF2 | 22155 | 877 | 1.362 | 0.0343 | No | ||
| 3 | CFL1 | 4516 | 1016 | 1.253 | 0.0551 | No | ||
| 4 | VDAC2 | 22081 | 1629 | 0.901 | 0.0423 | No | ||
| 5 | P4HA1 | 20017 | 1789 | 0.809 | 0.0519 | No | ||
| 6 | DDX3X | 24379 | 2114 | 0.640 | 0.0488 | No | ||
| 7 | ORC5L | 11173 3595 | 2295 | 0.544 | 0.0514 | No | ||
| 8 | DSCAM | 1667 4641 22530 | 2496 | 0.473 | 0.0512 | No | ||
| 9 | ZDHHC5 | 14547 | 2633 | 0.421 | 0.0533 | No | ||
| 10 | CDC42SE1 | 929 486 | 2729 | 0.384 | 0.0569 | No | ||
| 11 | ARCN1 | 19143 | 2834 | 0.354 | 0.0592 | No | ||
| 12 | ABR | 8468 20346 1476 1429 | 3185 | 0.269 | 0.0464 | No | ||
| 13 | CASK | 24184 2618 2604 | 3264 | 0.254 | 0.0479 | No | ||
| 14 | SPRYD3 | 522 | 3277 | 0.251 | 0.0529 | No | ||
| 15 | SLAMF6 | 6627 | 3313 | 0.244 | 0.0565 | No | ||
| 16 | DCLRE1B | 1925 8952 4714 | 4002 | 0.151 | 0.0227 | No | ||
| 17 | SMARCD1 | 2241 22360 2250 | 4486 | 0.112 | -0.0009 | No | ||
| 18 | TRIM44 | 8164 13487 | 4584 | 0.107 | -0.0037 | No | ||
| 19 | STAT3 | 5525 9906 | 4604 | 0.106 | -0.0023 | No | ||
| 20 | AQP4 | 4403 | 4642 | 0.103 | -0.0020 | No | ||
| 21 | TRIM33 | 8236 13579 15472 8237 | 4743 | 0.098 | -0.0052 | No | ||
| 22 | MUC4 | 8929 1746 | 4834 | 0.093 | -0.0080 | No | ||
| 23 | CTBP2 | 17591 1137 | 4842 | 0.092 | -0.0063 | No | ||
| 24 | CACNG1 | 20177 | 4912 | 0.090 | -0.0080 | No | ||
| 25 | TGFBR2 | 2994 3001 10167 | 5015 | 0.085 | -0.0116 | No | ||
| 26 | GABBR1 | 1519 7034 | 5132 | 0.080 | -0.0161 | No | ||
| 27 | DDX3Y | 6507 4593 | 5727 | 0.061 | -0.0468 | No | ||
| 28 | WDR26 | 13733 | 5767 | 0.060 | -0.0475 | No | ||
| 29 | ARID4B | 21705 3233 | 5891 | 0.057 | -0.0529 | No | ||
| 30 | CHRD | 22817 | 6399 | 0.045 | -0.0793 | No | ||
| 31 | NCDN | 15756 2487 6471 | 6890 | 0.036 | -0.1049 | No | ||
| 32 | KIAA0427 | 11228 | 6900 | 0.036 | -0.1046 | No | ||
| 33 | STX1B2 | 17609 | 6958 | 0.035 | -0.1069 | No | ||
| 34 | PTRF | 9667 | 7011 | 0.034 | -0.1089 | No | ||
| 35 | MTUS1 | 4162 158 | 7595 | 0.025 | -0.1399 | No | ||
| 36 | IXL | 17910 | 7910 | 0.021 | -0.1564 | No | ||
| 37 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 7968 | 0.020 | -0.1590 | No | ||
| 38 | PSD2 | 23597 | 8273 | 0.017 | -0.1750 | No | ||
| 39 | ST3GAL3 | 15782 | 8518 | 0.014 | -0.1879 | No | ||
| 40 | HSPA14 | 14695 | 8538 | 0.014 | -0.1886 | No | ||
| 41 | CLCA3 | 6224 | 8587 | 0.013 | -0.1909 | No | ||
| 42 | MTSS1 | 5585 22285 | 8675 | 0.012 | -0.1954 | No | ||
| 43 | GRIN3A | 15883 | 9176 | 0.007 | -0.2222 | No | ||
| 44 | SLC4A4 | 16807 12035 7036 | 9361 | 0.005 | -0.2321 | No | ||
| 45 | DNMT3A | 2167 21330 | 9546 | 0.003 | -0.2420 | No | ||
| 46 | PCDH10 | 5227 9534 1893 | 9959 | -0.002 | -0.2642 | No | ||
| 47 | NFASC | 11233 979 3952 4047 | 10013 | -0.003 | -0.2670 | No | ||
| 48 | CCNJ | 23859 | 10039 | -0.003 | -0.2683 | No | ||
| 49 | NFATC3 | 5169 | 10340 | -0.006 | -0.2843 | No | ||
| 50 | FBXO24 | 12918 3649 | 10363 | -0.006 | -0.2854 | No | ||
| 51 | ASB15 | 8129 | 10452 | -0.007 | -0.2900 | No | ||
| 52 | DMRTB1 | 15813 | 11380 | -0.017 | -0.3397 | No | ||
| 53 | E2F3 | 21500 8874 | 11388 | -0.017 | -0.3396 | No | ||
| 54 | MAP2K7 | 6453 | 11439 | -0.018 | -0.3419 | No | ||
| 55 | FGF7 | 8965 | 11966 | -0.025 | -0.3698 | No | ||
| 56 | DUSP3 | 1358 13022 | 12297 | -0.029 | -0.3870 | No | ||
| 57 | KLC2 | 23973 3701 | 12395 | -0.031 | -0.3915 | No | ||
| 58 | PLCG1 | 14753 | 12496 | -0.032 | -0.3962 | No | ||
| 59 | FRK | 20056 | 12656 | -0.035 | -0.4040 | No | ||
| 60 | FOSB | 17945 | 12846 | -0.038 | -0.4134 | No | ||
| 61 | CCNK | 21160 | 12849 | -0.038 | -0.4126 | No | ||
| 62 | ATXN1 | 21477 | 13172 | -0.044 | -0.4290 | No | ||
| 63 | NLGN2 | 10117 | 13313 | -0.047 | -0.4356 | No | ||
| 64 | TMEM47 | 5315 | 13692 | -0.056 | -0.4547 | No | ||
| 65 | SLC38A3 | 13319 | 13798 | -0.059 | -0.4591 | No | ||
| 66 | BAAT | 15885 | 13924 | -0.062 | -0.4644 | No | ||
| 67 | KIFC2 | 9226 22442 | 14310 | -0.074 | -0.4836 | No | ||
| 68 | KLF12 | 4960 2637 21732 9228 | 14352 | -0.076 | -0.4841 | No | ||
| 69 | TRAIP | 19315 | 14416 | -0.078 | -0.4857 | No | ||
| 70 | NCALD | 22310 | 14437 | -0.079 | -0.4850 | No | ||
| 71 | CNTNAP1 | 11987 | 15187 | -0.126 | -0.5227 | No | ||
| 72 | BAZ2A | 4371 | 15348 | -0.141 | -0.5281 | No | ||
| 73 | ANK3 | 8591 3445 3304 3381 | 15418 | -0.149 | -0.5285 | No | ||
| 74 | PKNOX2 | 3090 5511 9889 | 15510 | -0.159 | -0.5298 | No | ||
| 75 | SENP5 | 6737 11508 | 15626 | -0.177 | -0.5320 | No | ||
| 76 | TRIT1 | 16098 | 15828 | -0.215 | -0.5381 | Yes | ||
| 77 | RAP1GDS1 | 10487 | 15831 | -0.215 | -0.5333 | Yes | ||
| 78 | LIN28 | 15723 | 15884 | -0.225 | -0.5311 | Yes | ||
| 79 | SOX12 | 5478 | 15908 | -0.229 | -0.5271 | Yes | ||
| 80 | TPM4 | 11589 11588 6819 | 15909 | -0.230 | -0.5220 | Yes | ||
| 81 | PARP6 | 19419 2979 3156 | 15920 | -0.233 | -0.5172 | Yes | ||
| 82 | ARF1 | 64 | 15967 | -0.245 | -0.5142 | Yes | ||
| 83 | ING3 | 17514 1019 | 16093 | -0.275 | -0.5147 | Yes | ||
| 84 | HPS1 | 23671 | 16132 | -0.284 | -0.5104 | Yes | ||
| 85 | PPM1B | 5280 1567 5281 | 16297 | -0.332 | -0.5118 | Yes | ||
| 86 | USP37 | 13923 | 16324 | -0.341 | -0.5055 | Yes | ||
| 87 | BRCA1 | 20213 | 16654 | -0.466 | -0.5128 | Yes | ||
| 88 | TAP2 | 23282 | 16656 | -0.466 | -0.5023 | Yes | ||
| 89 | FBXW5 | 11393 | 16771 | -0.523 | -0.4967 | Yes | ||
| 90 | MAPRE1 | 4652 | 16854 | -0.563 | -0.4884 | Yes | ||
| 91 | DND1 | 10017 | 16858 | -0.566 | -0.4758 | Yes | ||
| 92 | MAP3K8 | 23495 | 17035 | -0.667 | -0.4703 | Yes | ||
| 93 | MARCKS | 9331 | 17217 | -0.800 | -0.4621 | Yes | ||
| 94 | NARG1 | 7934 917 | 17319 | -0.884 | -0.4476 | Yes | ||
| 95 | FOXP4 | 22949 | 17349 | -0.904 | -0.4288 | Yes | ||
| 96 | ZMYND11 | 12364 3269 3286 3223 | 17389 | -0.929 | -0.4100 | Yes | ||
| 97 | ABCB9 | 3576 3546 7096 | 17478 | -0.994 | -0.3923 | Yes | ||
| 98 | DNMT3B | 2840 14785 | 17503 | -1.004 | -0.3710 | Yes | ||
| 99 | PPIL2 | 22648 | 17529 | -1.018 | -0.3494 | Yes | ||
| 100 | CIT | 16727 | 17530 | -1.019 | -0.3264 | Yes | ||
| 101 | RAB3IP | 19623 | 17663 | -1.140 | -0.3078 | Yes | ||
| 102 | HNRPAB | 9104 | 17686 | -1.173 | -0.2826 | Yes | ||
| 103 | ILF3 | 3110 3030 9176 | 17724 | -1.199 | -0.2576 | Yes | ||
| 104 | OGT | 4241 24274 | 17835 | -1.316 | -0.2339 | Yes | ||
| 105 | CHST3 | 19760 | 17859 | -1.337 | -0.2050 | Yes | ||
| 106 | PDCD11 | 23834 | 18133 | -1.671 | -0.1821 | Yes | ||
| 107 | FBS1 | 18067 | 18153 | -1.696 | -0.1449 | Yes | ||
| 108 | ACACA | 309 4209 | 18306 | -2.010 | -0.1078 | Yes | ||
| 109 | PCTK1 | 24369 | 18357 | -2.140 | -0.0623 | Yes | ||
| 110 | UBAP2 | 15912 | 18571 | -3.385 | 0.0024 | Yes |