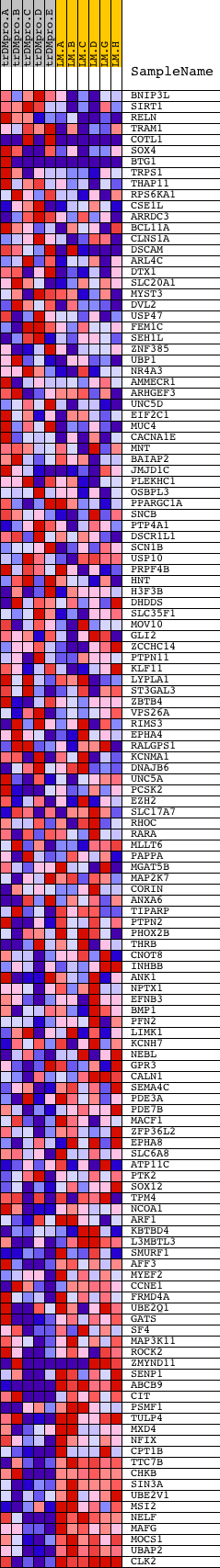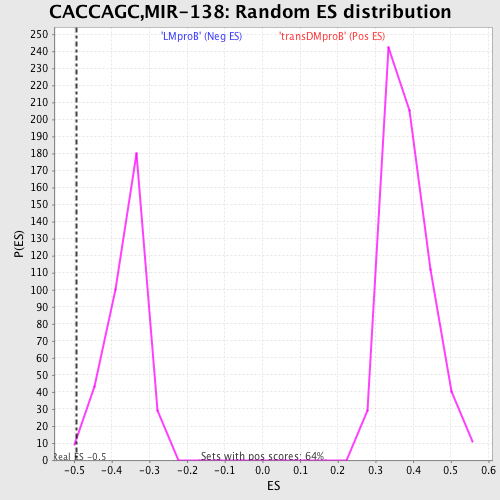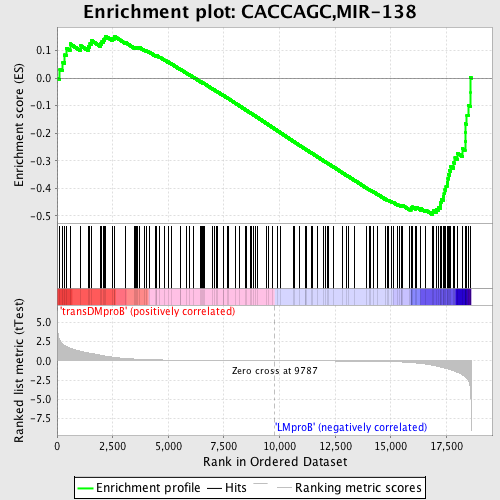
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | CACCAGC,MIR-138 |
| Enrichment Score (ES) | -0.49472982 |
| Normalized Enrichment Score (NES) | -1.3651047 |
| Nominal p-value | 0.008310249 |
| FDR q-value | 0.53890574 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BNIP3L | 21775 | 125 | 2.591 | 0.0330 | No | ||
| 2 | SIRT1 | 19745 | 258 | 2.110 | 0.0583 | No | ||
| 3 | RELN | 5372 | 341 | 1.962 | 0.0840 | No | ||
| 4 | TRAM1 | 13016 14000 | 411 | 1.841 | 0.1085 | No | ||
| 5 | COTL1 | 12987 7781 | 585 | 1.633 | 0.1243 | No | ||
| 6 | SOX4 | 5479 | 1064 | 1.224 | 0.1172 | No | ||
| 7 | BTG1 | 4458 4457 8663 | 1409 | 1.001 | 0.1140 | No | ||
| 8 | TRPS1 | 8195 | 1475 | 0.982 | 0.1255 | No | ||
| 9 | THAP11 | 18487 | 1536 | 0.951 | 0.1369 | No | ||
| 10 | RPS6KA1 | 15725 | 1965 | 0.709 | 0.1246 | No | ||
| 11 | CSE1L | 2906 | 2013 | 0.684 | 0.1326 | No | ||
| 12 | ARRDC3 | 4191 | 2084 | 0.651 | 0.1388 | No | ||
| 13 | BCL11A | 4691 | 2149 | 0.616 | 0.1448 | No | ||
| 14 | CLNS1A | 4526 4525 | 2180 | 0.596 | 0.1523 | No | ||
| 15 | DSCAM | 1667 4641 22530 | 2496 | 0.473 | 0.1426 | No | ||
| 16 | ARL4C | 13888 | 2562 | 0.444 | 0.1459 | No | ||
| 17 | DTX1 | 16396 | 2582 | 0.435 | 0.1515 | No | ||
| 18 | SLC20A1 | 14855 | 3055 | 0.298 | 0.1306 | No | ||
| 19 | MYST3 | 18651 | 3498 | 0.214 | 0.1099 | No | ||
| 20 | DVL2 | 20813 | 3517 | 0.211 | 0.1122 | No | ||
| 21 | USP47 | 6794 | 3585 | 0.201 | 0.1117 | No | ||
| 22 | FEM1C | 23441 10771 23442 | 3622 | 0.196 | 0.1127 | No | ||
| 23 | SEH1L | 13001 | 3721 | 0.185 | 0.1103 | No | ||
| 24 | ZNF385 | 22106 2297 | 3921 | 0.159 | 0.1020 | No | ||
| 25 | UBP1 | 5827 | 4024 | 0.149 | 0.0987 | No | ||
| 26 | NR4A3 | 9473 16212 5183 | 4142 | 0.138 | 0.0945 | No | ||
| 27 | AMMECR1 | 7067 | 4428 | 0.116 | 0.0809 | No | ||
| 28 | ARHGEF3 | 12952 | 4480 | 0.113 | 0.0799 | No | ||
| 29 | UNC5D | 9980 | 4483 | 0.112 | 0.0815 | No | ||
| 30 | EIF2C1 | 10672 | 4619 | 0.105 | 0.0758 | No | ||
| 31 | MUC4 | 8929 1746 | 4834 | 0.093 | 0.0656 | No | ||
| 32 | CACNA1E | 4465 | 4985 | 0.086 | 0.0588 | No | ||
| 33 | MNT | 20784 | 5133 | 0.080 | 0.0521 | No | ||
| 34 | BAIAP2 | 449 4236 | 5548 | 0.066 | 0.0307 | No | ||
| 35 | JMJD1C | 11531 19996 | 5556 | 0.066 | 0.0314 | No | ||
| 36 | PLEKHC1 | 21864 | 5816 | 0.059 | 0.0183 | No | ||
| 37 | OSBPL3 | 7747 998 | 5946 | 0.056 | 0.0121 | No | ||
| 38 | PPARGC1A | 16533 | 6137 | 0.051 | 0.0026 | No | ||
| 39 | SNCB | 21456 | 6441 | 0.044 | -0.0131 | No | ||
| 40 | PTP4A1 | 9658 | 6467 | 0.044 | -0.0138 | No | ||
| 41 | DSCR1L1 | 23231 | 6554 | 0.042 | -0.0178 | No | ||
| 42 | SCN1B | 1148 17872 | 6564 | 0.042 | -0.0176 | No | ||
| 43 | USP10 | 10248 3795 | 6612 | 0.041 | -0.0195 | No | ||
| 44 | PRPF4B | 3208 5295 | 6998 | 0.034 | -0.0398 | No | ||
| 45 | HNT | 10641 | 7063 | 0.033 | -0.0428 | No | ||
| 46 | H3F3B | 4839 | 7177 | 0.032 | -0.0484 | No | ||
| 47 | DHDDS | 2536 15724 | 7197 | 0.031 | -0.0489 | No | ||
| 48 | SLC35F1 | 20027 | 7472 | 0.027 | -0.0633 | No | ||
| 49 | MOV10 | 9403 9402 5114 | 7488 | 0.027 | -0.0637 | No | ||
| 50 | GLI2 | 13859 | 7660 | 0.024 | -0.0726 | No | ||
| 51 | ZCCHC14 | 4731 | 7708 | 0.024 | -0.0748 | No | ||
| 52 | PTPN11 | 5326 16391 9660 | 8010 | 0.020 | -0.0908 | No | ||
| 53 | KLF11 | 9700 | 8180 | 0.018 | -0.0997 | No | ||
| 54 | LYPLA1 | 4107 9580 | 8453 | 0.014 | -0.1142 | No | ||
| 55 | ST3GAL3 | 15782 | 8518 | 0.014 | -0.1174 | No | ||
| 56 | ZBTB4 | 13267 | 8682 | 0.012 | -0.1260 | No | ||
| 57 | VPS26A | 19752 3333 | 8750 | 0.011 | -0.1295 | No | ||
| 58 | RIMS3 | 6325 | 8806 | 0.011 | -0.1323 | No | ||
| 59 | EPHA4 | 4673 | 8930 | 0.009 | -0.1388 | No | ||
| 60 | RALGPS1 | 2672 6303 | 9002 | 0.008 | -0.1425 | No | ||
| 61 | KCNMA1 | 4947 | 9393 | 0.004 | -0.1636 | No | ||
| 62 | DNAJB6 | 6246 3538 | 9505 | 0.003 | -0.1695 | No | ||
| 63 | UNC5A | 3288 21631 | 9673 | 0.001 | -0.1785 | No | ||
| 64 | PCSK2 | 9537 5231 14824 | 9916 | -0.002 | -0.1916 | No | ||
| 65 | EZH2 | 1092 17163 | 10050 | -0.003 | -0.1987 | No | ||
| 66 | SLC17A7 | 13081 | 10621 | -0.009 | -0.2294 | No | ||
| 67 | RHOC | 15467 | 10677 | -0.009 | -0.2323 | No | ||
| 68 | RARA | 5358 | 10892 | -0.012 | -0.2437 | No | ||
| 69 | MLLT6 | 20678 | 11145 | -0.015 | -0.2571 | No | ||
| 70 | PAPPA | 16190 | 11171 | -0.015 | -0.2582 | No | ||
| 71 | MGAT5B | 20584 11197 | 11209 | -0.015 | -0.2600 | No | ||
| 72 | MAP2K7 | 6453 | 11439 | -0.018 | -0.2721 | No | ||
| 73 | CORIN | 16517 | 11495 | -0.019 | -0.2748 | No | ||
| 74 | ANXA6 | 20449 | 11698 | -0.022 | -0.2854 | No | ||
| 75 | TIPARP | 15576 | 11976 | -0.025 | -0.3000 | No | ||
| 76 | PTPN2 | 23414 | 12047 | -0.026 | -0.3034 | No | ||
| 77 | PHOX2B | 16524 | 12143 | -0.027 | -0.3081 | No | ||
| 78 | THRB | 10173 | 12192 | -0.028 | -0.3103 | No | ||
| 79 | CNOT8 | 12778 | 12427 | -0.031 | -0.3224 | No | ||
| 80 | INHBB | 13858 | 12808 | -0.037 | -0.3424 | No | ||
| 81 | ANK1 | 22501 18650 4385 8590 18649 | 13025 | -0.041 | -0.3535 | No | ||
| 82 | NPTX1 | 20126 | 13076 | -0.042 | -0.3555 | No | ||
| 83 | EFNB3 | 20391 | 13374 | -0.048 | -0.3709 | No | ||
| 84 | BMP1 | 3268 8658 21758 | 13927 | -0.062 | -0.3998 | No | ||
| 85 | PFN2 | 15337 1818 | 14042 | -0.065 | -0.4050 | No | ||
| 86 | LIMK1 | 16350 | 14087 | -0.067 | -0.4063 | No | ||
| 87 | KCNH7 | 2891 | 14223 | -0.071 | -0.4125 | No | ||
| 88 | NEBL | 14678 2691 | 14397 | -0.078 | -0.4207 | No | ||
| 89 | GPR3 | 15729 | 14750 | -0.093 | -0.4383 | No | ||
| 90 | CALN1 | 4713 | 14854 | -0.100 | -0.4423 | No | ||
| 91 | SEMA4C | 9799 | 14907 | -0.104 | -0.4436 | No | ||
| 92 | PDE3A | 7045 | 15024 | -0.112 | -0.4481 | No | ||
| 93 | PDE7B | 19807 | 15113 | -0.119 | -0.4510 | No | ||
| 94 | MACF1 | 4316 15763 2382 15764 | 15296 | -0.136 | -0.4588 | No | ||
| 95 | ZFP36L2 | 8662 | 15387 | -0.145 | -0.4614 | No | ||
| 96 | EPHA8 | 15705 | 15496 | -0.158 | -0.4649 | No | ||
| 97 | SLC6A8 | 24306 | 15501 | -0.158 | -0.4626 | No | ||
| 98 | ATP11C | 6813 24146 | 15546 | -0.164 | -0.4625 | No | ||
| 99 | PTK2 | 22271 | 15843 | -0.218 | -0.4752 | No | ||
| 100 | SOX12 | 5478 | 15908 | -0.229 | -0.4751 | No | ||
| 101 | TPM4 | 11589 11588 6819 | 15909 | -0.230 | -0.4716 | No | ||
| 102 | NCOA1 | 5154 | 15919 | -0.233 | -0.4685 | No | ||
| 103 | ARF1 | 64 | 15967 | -0.245 | -0.4673 | No | ||
| 104 | KBTBD4 | 2723 14952 | 16116 | -0.281 | -0.4710 | No | ||
| 105 | L3MBTL3 | 6195 3411 | 16140 | -0.289 | -0.4678 | No | ||
| 106 | SMURF1 | 13289 7989 | 16354 | -0.352 | -0.4739 | No | ||
| 107 | AFF3 | 4982 | 16570 | -0.429 | -0.4789 | No | ||
| 108 | MYEF2 | 5140 | 16863 | -0.567 | -0.4860 | Yes | ||
| 109 | CCNE1 | 17857 | 16907 | -0.589 | -0.4793 | Yes | ||
| 110 | FRMD4A | 5557 | 17055 | -0.679 | -0.4768 | Yes | ||
| 111 | UBE2Q1 | 13662 12851 15537 | 17135 | -0.732 | -0.4698 | Yes | ||
| 112 | GATS | 16683 | 17224 | -0.805 | -0.4622 | Yes | ||
| 113 | SF4 | 18593 | 17247 | -0.823 | -0.4508 | Yes | ||
| 114 | MAP3K11 | 11163 | 17263 | -0.835 | -0.4388 | Yes | ||
| 115 | ROCK2 | 21309 5387 | 17355 | -0.908 | -0.4297 | Yes | ||
| 116 | ZMYND11 | 12364 3269 3286 3223 | 17389 | -0.929 | -0.4173 | Yes | ||
| 117 | SENP1 | 10301 | 17432 | -0.959 | -0.4048 | Yes | ||
| 118 | ABCB9 | 3576 3546 7096 | 17478 | -0.994 | -0.3920 | Yes | ||
| 119 | CIT | 16727 | 17530 | -1.019 | -0.3791 | Yes | ||
| 120 | PSMF1 | 6009 | 17566 | -1.048 | -0.3649 | Yes | ||
| 121 | TULP4 | 12740 | 17591 | -1.066 | -0.3498 | Yes | ||
| 122 | MXD4 | 9346 | 17634 | -1.101 | -0.3351 | Yes | ||
| 123 | NFIX | 9458 | 17695 | -1.179 | -0.3203 | Yes | ||
| 124 | CPT1B | 2187 22159 | 17831 | -1.312 | -0.3074 | Yes | ||
| 125 | TTC7B | 21006 | 17882 | -1.357 | -0.2893 | Yes | ||
| 126 | CHKB | 4518 | 17993 | -1.498 | -0.2722 | Yes | ||
| 127 | SIN3A | 5442 | 18231 | -1.862 | -0.2564 | Yes | ||
| 128 | UBE2V1 | 12381 | 18338 | -2.103 | -0.2299 | Yes | ||
| 129 | MSI2 | 8030 1268 | 18347 | -2.118 | -0.1978 | Yes | ||
| 130 | NELF | 12174 | 18354 | -2.138 | -0.1653 | Yes | ||
| 131 | MAFG | 5060 | 18420 | -2.276 | -0.1338 | Yes | ||
| 132 | MOCS1 | 7151 | 18479 | -2.535 | -0.0980 | Yes | ||
| 133 | UBAP2 | 15912 | 18571 | -3.385 | -0.0510 | Yes | ||
| 134 | CLK2 | 1883 15544 | 18575 | -3.473 | 0.0022 | Yes |