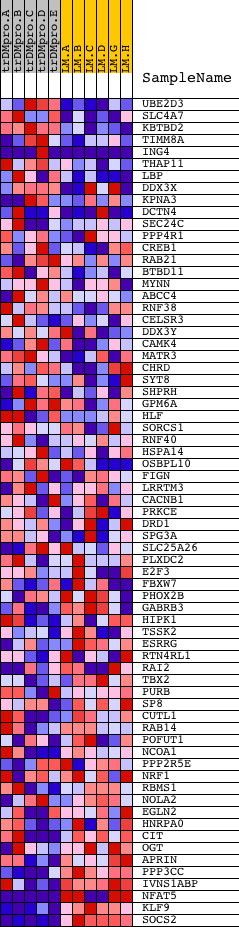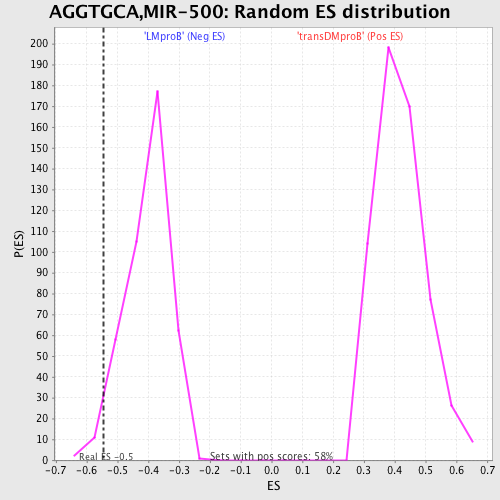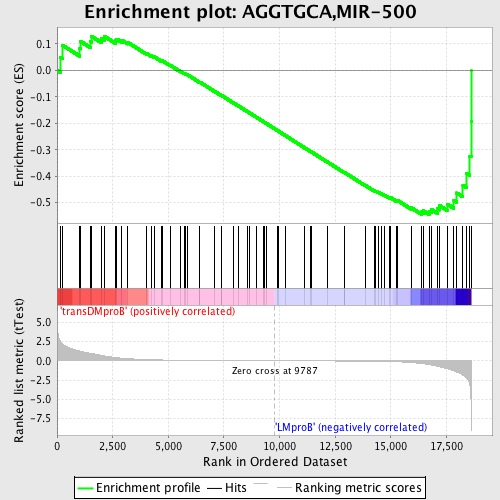
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | AGGTGCA,MIR-500 |
| Enrichment Score (ES) | -0.5446716 |
| Normalized Enrichment Score (NES) | -1.3586985 |
| Nominal p-value | 0.026442308 |
| FDR q-value | 0.5375804 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | UBE2D3 | 7253 | 153 | 2.446 | 0.0492 | No | ||
| 2 | SLC4A7 | 10181 | 243 | 2.153 | 0.0950 | No | ||
| 3 | KBTBD2 | 17137 | 1020 | 1.251 | 0.0826 | No | ||
| 4 | TIMM8A | 24062 | 1059 | 1.226 | 0.1094 | No | ||
| 5 | ING4 | 6602 1155 1102 11371 | 1499 | 0.972 | 0.1085 | No | ||
| 6 | THAP11 | 18487 | 1536 | 0.951 | 0.1289 | No | ||
| 7 | LBP | 14760 | 1990 | 0.694 | 0.1208 | No | ||
| 8 | DDX3X | 24379 | 2114 | 0.640 | 0.1293 | No | ||
| 9 | KPNA3 | 21797 | 2621 | 0.424 | 0.1119 | No | ||
| 10 | DCTN4 | 12560 7474 | 2681 | 0.401 | 0.1182 | No | ||
| 11 | SEC24C | 22086 | 2910 | 0.335 | 0.1138 | No | ||
| 12 | PPP4R1 | 7670 7669 | 3176 | 0.270 | 0.1058 | No | ||
| 13 | CREB1 | 3990 8782 4558 4093 | 4019 | 0.150 | 0.0639 | No | ||
| 14 | RAB21 | 5696 | 4225 | 0.130 | 0.0559 | No | ||
| 15 | BTBD11 | 13147 | 4366 | 0.119 | 0.0512 | No | ||
| 16 | MYNN | 15620 1914 | 4705 | 0.100 | 0.0353 | No | ||
| 17 | ABCC4 | 21721 | 4723 | 0.099 | 0.0367 | No | ||
| 18 | RNF38 | 7862 2328 13115 | 5085 | 0.082 | 0.0192 | No | ||
| 19 | CELSR3 | 19305 8419 | 5561 | 0.066 | -0.0049 | No | ||
| 20 | DDX3Y | 6507 4593 | 5727 | 0.061 | -0.0123 | No | ||
| 21 | CAMK4 | 4473 | 5783 | 0.060 | -0.0139 | No | ||
| 22 | MATR3 | 2043 5081 | 5845 | 0.058 | -0.0158 | No | ||
| 23 | CHRD | 22817 | 6399 | 0.045 | -0.0446 | No | ||
| 24 | SYT8 | 18006 | 6412 | 0.045 | -0.0441 | No | ||
| 25 | SHPRH | 6477 | 7057 | 0.033 | -0.0781 | No | ||
| 26 | GPM6A | 3834 10618 | 7382 | 0.028 | -0.0949 | No | ||
| 27 | HLF | 10124 | 7395 | 0.028 | -0.0949 | No | ||
| 28 | SORCS1 | 7184 12203 | 7908 | 0.021 | -0.1220 | No | ||
| 29 | RNF40 | 6133 | 8150 | 0.018 | -0.1346 | No | ||
| 30 | HSPA14 | 14695 | 8538 | 0.014 | -0.1551 | No | ||
| 31 | OSBPL10 | 13213 | 8635 | 0.013 | -0.1600 | No | ||
| 32 | FIGN | 14575 | 8983 | 0.009 | -0.1785 | No | ||
| 33 | LRRTM3 | 19744 | 9292 | 0.005 | -0.1950 | No | ||
| 34 | CACNB1 | 1443 1304 | 9319 | 0.005 | -0.1962 | No | ||
| 35 | PRKCE | 9575 | 9432 | 0.004 | -0.2022 | No | ||
| 36 | DRD1 | 4638 | 9892 | -0.001 | -0.2269 | No | ||
| 37 | SPG3A | 7878 13145 | 9971 | -0.002 | -0.2311 | No | ||
| 38 | SLC25A26 | 12552 | 10279 | -0.005 | -0.2475 | No | ||
| 39 | PLXDC2 | 15114 | 11137 | -0.015 | -0.2933 | No | ||
| 40 | E2F3 | 21500 8874 | 11388 | -0.017 | -0.3064 | No | ||
| 41 | FBXW7 | 1805 11928 | 11412 | -0.018 | -0.3072 | No | ||
| 42 | PHOX2B | 16524 | 12143 | -0.027 | -0.3460 | No | ||
| 43 | GABRB3 | 4747 | 12896 | -0.039 | -0.3856 | No | ||
| 44 | HIPK1 | 4851 | 12922 | -0.039 | -0.3860 | No | ||
| 45 | TSSK2 | 10225 | 13861 | -0.060 | -0.4352 | No | ||
| 46 | ESRRG | 6447 | 14257 | -0.072 | -0.4548 | No | ||
| 47 | RTN4RL1 | 20782 | 14327 | -0.075 | -0.4567 | No | ||
| 48 | RAI2 | 10762 | 14446 | -0.079 | -0.4612 | No | ||
| 49 | TBX2 | 20720 | 14571 | -0.084 | -0.4659 | No | ||
| 50 | PURB | 5335 | 14734 | -0.092 | -0.4725 | No | ||
| 51 | SP8 | 21122 | 14956 | -0.107 | -0.4819 | No | ||
| 52 | CUTL1 | 3507 3480 8820 3609 4577 | 14989 | -0.110 | -0.4810 | No | ||
| 53 | RAB14 | 12685 | 15275 | -0.134 | -0.4933 | No | ||
| 54 | POFUT1 | 4697 | 15290 | -0.135 | -0.4909 | No | ||
| 55 | NCOA1 | 5154 | 15919 | -0.233 | -0.5192 | No | ||
| 56 | PPP2R5E | 6523 2136 | 16368 | -0.358 | -0.5350 | Yes | ||
| 57 | NRF1 | 17499 | 16458 | -0.393 | -0.5305 | Yes | ||
| 58 | RBMS1 | 14580 | 16721 | -0.499 | -0.5330 | Yes | ||
| 59 | NOLA2 | 11972 | 16820 | -0.543 | -0.5255 | Yes | ||
| 60 | EGLN2 | 17920 | 17088 | -0.704 | -0.5233 | Yes | ||
| 61 | HNRPA0 | 13382 | 17172 | -0.759 | -0.5100 | Yes | ||
| 62 | CIT | 16727 | 17530 | -1.019 | -0.5053 | Yes | ||
| 63 | OGT | 4241 24274 | 17835 | -1.316 | -0.4907 | Yes | ||
| 64 | APRIN | 4145 | 17968 | -1.465 | -0.4634 | Yes | ||
| 65 | PPP3CC | 21763 | 18241 | -1.882 | -0.4339 | Yes | ||
| 66 | IVNS1ABP | 4384 4050 | 18414 | -2.264 | -0.3899 | Yes | ||
| 67 | NFAT5 | 3921 7037 12036 | 18552 | -3.129 | -0.3238 | Yes | ||
| 68 | KLF9 | 23903 | 18608 | -5.627 | -0.1945 | Yes | ||
| 69 | SOCS2 | 5694 | 18614 | -8.294 | 0.0001 | Yes |