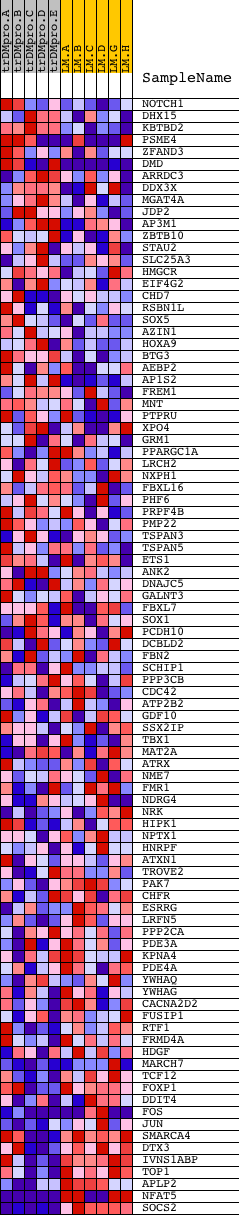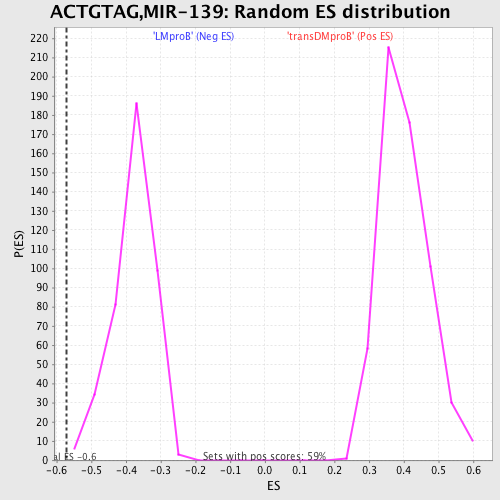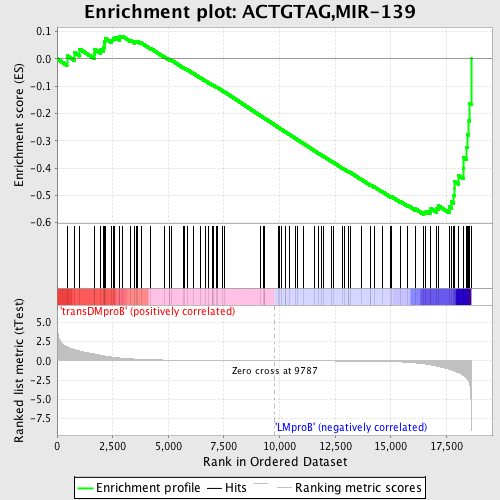
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | ACTGTAG,MIR-139 |
| Enrichment Score (ES) | -0.5701414 |
| Normalized Enrichment Score (NES) | -1.5068759 |
| Nominal p-value | 0.002444988 |
| FDR q-value | 0.6444573 |
| FWER p-Value | 0.797 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NOTCH1 | 14649 | 445 | 1.788 | 0.0122 | No | ||
| 2 | DHX15 | 8842 | 794 | 1.437 | 0.0225 | No | ||
| 3 | KBTBD2 | 17137 | 1020 | 1.251 | 0.0357 | No | ||
| 4 | PSME4 | 8339 20934 | 1676 | 0.878 | 0.0181 | No | ||
| 5 | ZFAND3 | 23306 | 1696 | 0.862 | 0.0346 | No | ||
| 6 | DMD | 24295 2647 | 1963 | 0.710 | 0.0346 | No | ||
| 7 | ARRDC3 | 4191 | 2084 | 0.651 | 0.0413 | No | ||
| 8 | DDX3X | 24379 | 2114 | 0.640 | 0.0527 | No | ||
| 9 | MGAT4A | 13977 | 2139 | 0.622 | 0.0640 | No | ||
| 10 | JDP2 | 21201 | 2157 | 0.612 | 0.0755 | No | ||
| 11 | AP3M1 | 7059 | 2460 | 0.484 | 0.0690 | No | ||
| 12 | ZBTB10 | 10468 | 2515 | 0.464 | 0.0754 | No | ||
| 13 | STAU2 | 6615 | 2599 | 0.429 | 0.0796 | No | ||
| 14 | SLC25A3 | 19644 | 2818 | 0.359 | 0.0751 | No | ||
| 15 | HMGCR | 9097 | 2819 | 0.359 | 0.0824 | No | ||
| 16 | EIF4G2 | 1908 8892 | 2927 | 0.331 | 0.0833 | No | ||
| 17 | CHD7 | 16278 | 3311 | 0.244 | 0.0676 | No | ||
| 18 | RSBN1L | 16598 | 3494 | 0.215 | 0.0621 | No | ||
| 19 | SOX5 | 9850 1044 5480 16937 | 3554 | 0.205 | 0.0631 | No | ||
| 20 | AZIN1 | 22307 | 3597 | 0.199 | 0.0648 | No | ||
| 21 | HOXA9 | 17147 9109 1024 | 3775 | 0.179 | 0.0589 | No | ||
| 22 | BTG3 | 8664 | 4217 | 0.131 | 0.0378 | No | ||
| 23 | AEBP2 | 4359 | 4823 | 0.093 | 0.0070 | No | ||
| 24 | AP1S2 | 8423 4228 | 5046 | 0.084 | -0.0033 | No | ||
| 25 | FREM1 | 141 | 5050 | 0.083 | -0.0018 | No | ||
| 26 | MNT | 20784 | 5133 | 0.080 | -0.0046 | No | ||
| 27 | PTPRU | 15738 | 5665 | 0.063 | -0.0320 | No | ||
| 28 | XPO4 | 7161 | 5739 | 0.061 | -0.0347 | No | ||
| 29 | GRM1 | 19822 | 5878 | 0.057 | -0.0410 | No | ||
| 30 | PPARGC1A | 16533 | 6137 | 0.051 | -0.0539 | No | ||
| 31 | LRCH2 | 24035 | 6429 | 0.045 | -0.0687 | No | ||
| 32 | NXPH1 | 17524 | 6679 | 0.040 | -0.0813 | No | ||
| 33 | FBXL16 | 10071 | 6823 | 0.037 | -0.0883 | No | ||
| 34 | PHF6 | 2648 24340 | 6970 | 0.035 | -0.0955 | No | ||
| 35 | PRPF4B | 3208 5295 | 6998 | 0.034 | -0.0962 | No | ||
| 36 | PMP22 | 9594 1312 | 7041 | 0.034 | -0.0978 | No | ||
| 37 | TSPAN3 | 3132 19105 | 7176 | 0.032 | -0.1044 | No | ||
| 38 | TSPAN5 | 15402 | 7208 | 0.031 | -0.1054 | No | ||
| 39 | ETS1 | 10715 6230 3135 | 7415 | 0.028 | -0.1160 | No | ||
| 40 | ANK2 | 4275 | 7516 | 0.026 | -0.1208 | No | ||
| 41 | DNAJC5 | 4568 | 9131 | 0.007 | -0.2078 | No | ||
| 42 | GALNT3 | 4751 | 9284 | 0.005 | -0.2159 | No | ||
| 43 | FBXL7 | 6691 | 9326 | 0.005 | -0.2180 | No | ||
| 44 | SOX1 | 18685 | 9335 | 0.005 | -0.2184 | No | ||
| 45 | PCDH10 | 5227 9534 1893 | 9959 | -0.002 | -0.2520 | No | ||
| 46 | DCBLD2 | 7855 | 10000 | -0.003 | -0.2541 | No | ||
| 47 | FBN2 | 23433 | 10071 | -0.003 | -0.2578 | No | ||
| 48 | SCHIP1 | 15569 | 10261 | -0.005 | -0.2679 | No | ||
| 49 | PPP3CB | 5285 | 10275 | -0.005 | -0.2685 | No | ||
| 50 | CDC42 | 4503 8722 4504 2465 | 10448 | -0.007 | -0.2776 | No | ||
| 51 | ATP2B2 | 17038 | 10699 | -0.010 | -0.2909 | No | ||
| 52 | GDF10 | 4767 | 10819 | -0.011 | -0.2971 | No | ||
| 53 | SSX2IP | 13617 | 11059 | -0.014 | -0.3097 | No | ||
| 54 | TBX1 | 10039 5630 | 11558 | -0.019 | -0.3362 | No | ||
| 55 | MAT2A | 10553 10554 6099 | 11769 | -0.022 | -0.3471 | No | ||
| 56 | ATRX | 10350 5922 2628 | 11889 | -0.024 | -0.3530 | No | ||
| 57 | NME7 | 926 4101 14070 3986 3946 931 | 11951 | -0.025 | -0.3558 | No | ||
| 58 | FMR1 | 24321 | 12334 | -0.030 | -0.3759 | No | ||
| 59 | NDRG4 | 10626 | 12406 | -0.031 | -0.3791 | No | ||
| 60 | NRK | 6559 | 12827 | -0.038 | -0.4010 | No | ||
| 61 | HIPK1 | 4851 | 12922 | -0.039 | -0.4053 | No | ||
| 62 | NPTX1 | 20126 | 13076 | -0.042 | -0.4127 | No | ||
| 63 | HNRPF | 13607 | 13113 | -0.043 | -0.4137 | No | ||
| 64 | ATXN1 | 21477 | 13172 | -0.044 | -0.4160 | No | ||
| 65 | TROVE2 | 13812 | 13677 | -0.055 | -0.4421 | No | ||
| 66 | PAK7 | 14417 | 14073 | -0.066 | -0.4621 | No | ||
| 67 | CHFR | 3646 16757 3624 | 14105 | -0.067 | -0.4624 | No | ||
| 68 | ESRRG | 6447 | 14257 | -0.072 | -0.4691 | No | ||
| 69 | LRFN5 | 6213 | 14610 | -0.086 | -0.4863 | No | ||
| 70 | PPP2CA | 20890 | 15002 | -0.111 | -0.5052 | No | ||
| 71 | PDE3A | 7045 | 15024 | -0.112 | -0.5040 | No | ||
| 72 | KPNA4 | 15321 | 15435 | -0.151 | -0.5231 | No | ||
| 73 | PDE4A | 3037 19542 3083 3023 | 15755 | -0.201 | -0.5363 | No | ||
| 74 | YWHAQ | 10369 | 16094 | -0.276 | -0.5489 | No | ||
| 75 | YWHAG | 16339 | 16488 | -0.404 | -0.5620 | Yes | ||
| 76 | CACNA2D2 | 7154 | 16578 | -0.431 | -0.5580 | Yes | ||
| 77 | FUSIP1 | 4715 16036 | 16764 | -0.520 | -0.5575 | Yes | ||
| 78 | RTF1 | 14899 | 16800 | -0.535 | -0.5486 | Yes | ||
| 79 | FRMD4A | 5557 | 17055 | -0.679 | -0.5485 | Yes | ||
| 80 | HDGF | 9082 4846 4845 | 17144 | -0.738 | -0.5383 | Yes | ||
| 81 | MARCH7 | 7172 2698 | 17626 | -1.097 | -0.5421 | Yes | ||
| 82 | TCF12 | 10044 3125 | 17737 | -1.219 | -0.5233 | Yes | ||
| 83 | FOXP1 | 4242 | 17833 | -1.313 | -0.5019 | Yes | ||
| 84 | DDIT4 | 19762 | 17845 | -1.322 | -0.4757 | Yes | ||
| 85 | FOS | 21202 | 17848 | -1.325 | -0.4490 | Yes | ||
| 86 | JUN | 15832 | 18032 | -1.559 | -0.4273 | Yes | ||
| 87 | SMARCA4 | 19538 | 18261 | -1.921 | -0.4007 | Yes | ||
| 88 | DTX3 | 19607 | 18280 | -1.947 | -0.3622 | Yes | ||
| 89 | IVNS1ABP | 4384 4050 | 18414 | -2.264 | -0.3236 | Yes | ||
| 90 | TOP1 | 5790 5789 10210 | 18440 | -2.355 | -0.2772 | Yes | ||
| 91 | APLP2 | 19190 | 18511 | -2.726 | -0.2258 | Yes | ||
| 92 | NFAT5 | 3921 7037 12036 | 18552 | -3.129 | -0.1646 | Yes | ||
| 93 | SOCS2 | 5694 | 18614 | -8.294 | 0.0001 | Yes |