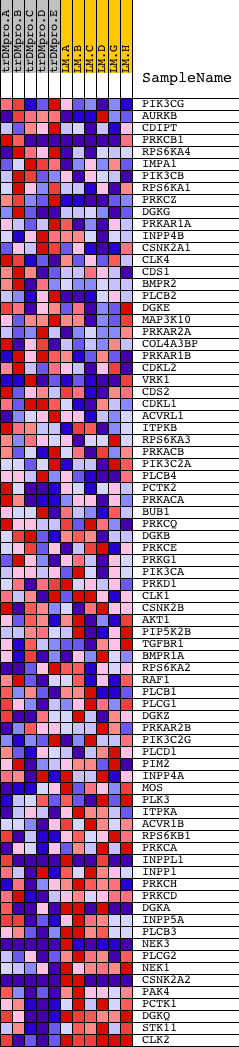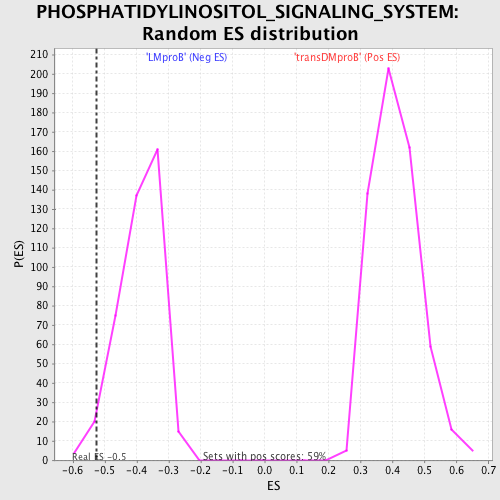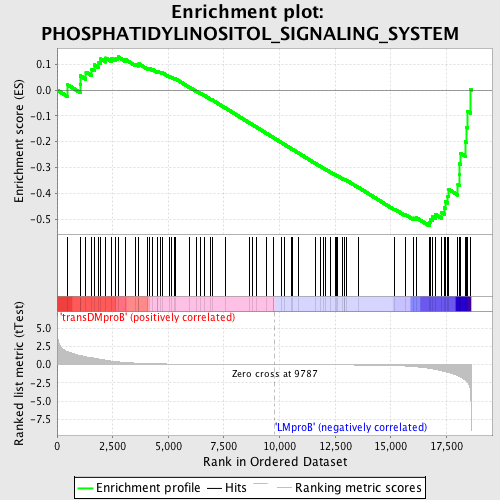
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | PHOSPHATIDYLINOSITOL_SIGNALING_SYSTEM |
| Enrichment Score (ES) | -0.5268411 |
| Normalized Enrichment Score (NES) | -1.3543863 |
| Nominal p-value | 0.024271844 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PIK3CG | 6635 | 476 | 1.747 | 0.0204 | No | ||
| 2 | AURKB | 20832 | 1035 | 1.245 | 0.0231 | No | ||
| 3 | CDIPT | 18071 | 1048 | 1.235 | 0.0550 | No | ||
| 4 | PRKCB1 | 1693 9574 | 1297 | 1.068 | 0.0698 | No | ||
| 5 | RPS6KA4 | 23804 | 1555 | 0.941 | 0.0807 | No | ||
| 6 | IMPA1 | 12066 | 1670 | 0.879 | 0.0978 | No | ||
| 7 | PIK3CB | 19030 | 1877 | 0.759 | 0.1067 | No | ||
| 8 | RPS6KA1 | 15725 | 1965 | 0.709 | 0.1207 | No | ||
| 9 | PRKCZ | 5260 | 2187 | 0.593 | 0.1244 | No | ||
| 10 | DGKG | 4282 | 2448 | 0.488 | 0.1232 | No | ||
| 11 | PRKAR1A | 20615 | 2643 | 0.415 | 0.1237 | No | ||
| 12 | INPP4B | 18557 | 2736 | 0.382 | 0.1288 | No | ||
| 13 | CSNK2A1 | 14797 | 3073 | 0.293 | 0.1184 | No | ||
| 14 | CLK4 | 20895 1292 | 3500 | 0.214 | 0.1011 | No | ||
| 15 | CDS1 | 16779 | 3648 | 0.193 | 0.0982 | No | ||
| 16 | BMPR2 | 14241 4081 | 3678 | 0.190 | 0.1017 | No | ||
| 17 | PLCB2 | 5262 | 4054 | 0.147 | 0.0853 | No | ||
| 18 | DGKE | 7069 | 4166 | 0.136 | 0.0829 | No | ||
| 19 | MAP3K10 | 3831 17916 | 4302 | 0.124 | 0.0789 | No | ||
| 20 | PRKAR2A | 5287 | 4503 | 0.111 | 0.0710 | No | ||
| 21 | COL4A3BP | 3221 7522 | 4504 | 0.111 | 0.0739 | No | ||
| 22 | PRKAR1B | 16320 | 4627 | 0.104 | 0.0701 | No | ||
| 23 | CDKL2 | 16482 3473 12001 3464 | 4731 | 0.099 | 0.0671 | No | ||
| 24 | VRK1 | 5862 2049 | 5043 | 0.084 | 0.0526 | No | ||
| 25 | CDS2 | 14837 | 5139 | 0.080 | 0.0495 | No | ||
| 26 | CDKL1 | 12914 | 5280 | 0.075 | 0.0440 | No | ||
| 27 | ACVRL1 | 22354 | 5322 | 0.074 | 0.0437 | No | ||
| 28 | ITPKB | 3963 14028 | 5972 | 0.055 | 0.0101 | No | ||
| 29 | RPS6KA3 | 8490 | 6252 | 0.048 | -0.0037 | No | ||
| 30 | PRKACB | 15140 | 6459 | 0.044 | -0.0136 | No | ||
| 31 | PIK3C2A | 17667 | 6629 | 0.041 | -0.0217 | No | ||
| 32 | PLCB4 | 9586 | 6893 | 0.036 | -0.0349 | No | ||
| 33 | PCTK2 | 19907 | 6966 | 0.035 | -0.0378 | No | ||
| 34 | PRKACA | 18549 3844 | 7558 | 0.026 | -0.0690 | No | ||
| 35 | BUB1 | 8665 | 8646 | 0.012 | -0.1274 | No | ||
| 36 | PRKCQ | 2873 2831 | 8796 | 0.011 | -0.1351 | No | ||
| 37 | DGKB | 5722 | 8950 | 0.009 | -0.1431 | No | ||
| 38 | PRKCE | 9575 | 9432 | 0.004 | -0.1690 | No | ||
| 39 | PRKG1 | 5289 | 9719 | 0.001 | -0.1844 | No | ||
| 40 | PIK3CA | 9562 | 9725 | 0.001 | -0.1846 | No | ||
| 41 | PRKD1 | 2074 21075 | 10092 | -0.003 | -0.2043 | No | ||
| 42 | CLK1 | 13951 4127 4529 8748 | 10212 | -0.005 | -0.2106 | No | ||
| 43 | CSNK2B | 23008 | 10542 | -0.008 | -0.2281 | No | ||
| 44 | AKT1 | 8568 | 10562 | -0.008 | -0.2289 | No | ||
| 45 | PIP5K2B | 20269 1254 | 10849 | -0.011 | -0.2440 | No | ||
| 46 | TGFBR1 | 16213 10165 5745 | 11615 | -0.020 | -0.2848 | No | ||
| 47 | BMPR1A | 8660 4450 | 11852 | -0.023 | -0.2969 | No | ||
| 48 | RPS6KA2 | 9759 9758 | 11961 | -0.025 | -0.3021 | No | ||
| 49 | RAF1 | 17035 | 12066 | -0.026 | -0.3070 | No | ||
| 50 | PLCB1 | 14832 2821 | 12286 | -0.029 | -0.3180 | No | ||
| 51 | PLCG1 | 14753 | 12496 | -0.032 | -0.3285 | No | ||
| 52 | DGKZ | 2836 14522 | 12567 | -0.033 | -0.3314 | No | ||
| 53 | PRKAR2B | 5288 2107 | 12603 | -0.034 | -0.3324 | No | ||
| 54 | PIK3C2G | 17253 | 12841 | -0.038 | -0.3442 | No | ||
| 55 | PLCD1 | 18972 | 12905 | -0.039 | -0.3465 | No | ||
| 56 | PIM2 | 5249 9564 9565 | 12913 | -0.039 | -0.3459 | No | ||
| 57 | INPP4A | 3985 3935 11237 4085 | 13028 | -0.041 | -0.3509 | No | ||
| 58 | MOS | 16254 | 13528 | -0.051 | -0.3765 | No | ||
| 59 | PLK3 | 15790 | 15168 | -0.124 | -0.4616 | No | ||
| 60 | ITPKA | 14898 | 15646 | -0.181 | -0.4826 | No | ||
| 61 | ACVR1B | 4335 22353 | 16019 | -0.256 | -0.4959 | No | ||
| 62 | RPS6KB1 | 7815 1207 13040 | 16151 | -0.292 | -0.4953 | No | ||
| 63 | PRKCA | 20174 | 16737 | -0.507 | -0.5135 | Yes | ||
| 64 | INPPL1 | 17733 3775 | 16769 | -0.521 | -0.5014 | Yes | ||
| 65 | INPP1 | 13961 | 16868 | -0.570 | -0.4917 | Yes | ||
| 66 | PRKCH | 21246 | 17001 | -0.647 | -0.4817 | Yes | ||
| 67 | PRKCD | 21897 | 17286 | -0.851 | -0.4746 | Yes | ||
| 68 | DGKA | 3359 19589 | 17411 | -0.947 | -0.4563 | Yes | ||
| 69 | INPP5A | 10003 | 17454 | -0.985 | -0.4326 | Yes | ||
| 70 | PLCB3 | 23799 | 17555 | -1.037 | -0.4107 | Yes | ||
| 71 | NEK3 | 3778 18916 | 17600 | -1.076 | -0.3847 | Yes | ||
| 72 | PLCG2 | 18453 | 18016 | -1.534 | -0.3667 | Yes | ||
| 73 | NEK1 | 18610 | 18072 | -1.587 | -0.3278 | Yes | ||
| 74 | CSNK2A2 | 4567 8808 | 18083 | -1.602 | -0.2861 | Yes | ||
| 75 | PAK4 | 17909 | 18131 | -1.667 | -0.2447 | Yes | ||
| 76 | PCTK1 | 24369 | 18357 | -2.140 | -0.2004 | Yes | ||
| 77 | DGKQ | 4287 | 18398 | -2.216 | -0.1442 | Yes | ||
| 78 | STK11 | 19950 | 18455 | -2.438 | -0.0829 | Yes | ||
| 79 | CLK2 | 1883 15544 | 18575 | -3.473 | 0.0022 | Yes |