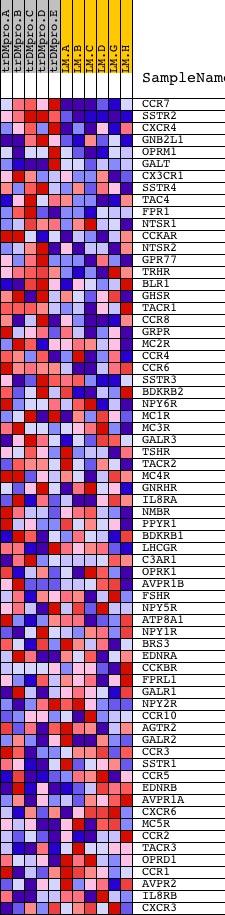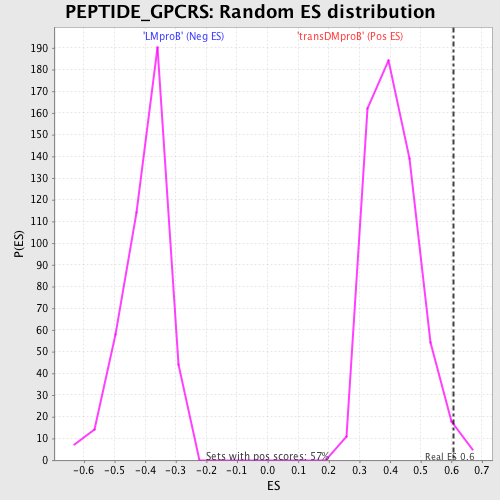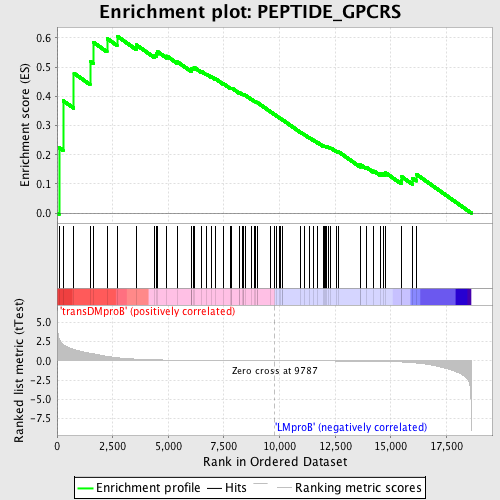
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | PEPTIDE_GPCRS |
| Enrichment Score (ES) | 0.60638446 |
| Normalized Enrichment Score (NES) | 1.4709412 |
| Nominal p-value | 0.015706806 |
| FDR q-value | 0.3531612 |
| FWER p-Value | 0.997 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CCR7 | 20254 | 88 | 2.803 | 0.2247 | Yes | ||
| 2 | SSTR2 | 20609 | 271 | 2.079 | 0.3851 | Yes | ||
| 3 | CXCR4 | 13844 | 758 | 1.469 | 0.4792 | Yes | ||
| 4 | GNB2L1 | 20911 | 1483 | 0.977 | 0.5201 | Yes | ||
| 5 | OPRM1 | 5212 | 1644 | 0.893 | 0.5846 | Yes | ||
| 6 | GALT | 16237 2399 | 2245 | 0.563 | 0.5983 | Yes | ||
| 7 | CX3CR1 | 18965 | 2696 | 0.395 | 0.6064 | Yes | ||
| 8 | SSTR4 | 9836 | 3561 | 0.203 | 0.5765 | No | ||
| 9 | TAC4 | 20696 | 4395 | 0.118 | 0.5412 | No | ||
| 10 | FPR1 | 23115 | 4465 | 0.114 | 0.5468 | No | ||
| 11 | NTSR1 | 14710 | 4500 | 0.112 | 0.5541 | No | ||
| 12 | CCKAR | 16531 | 4928 | 0.089 | 0.5383 | No | ||
| 13 | NTSR2 | 21310 | 5410 | 0.071 | 0.5182 | No | ||
| 14 | GPR77 | 17958 | 6047 | 0.053 | 0.4882 | No | ||
| 15 | TRHR | 22482 | 6048 | 0.053 | 0.4926 | No | ||
| 16 | BLR1 | 19145 3154 | 6050 | 0.053 | 0.4968 | No | ||
| 17 | GHSR | 1810 15625 | 6123 | 0.051 | 0.4972 | No | ||
| 18 | TACR1 | 1082 17403 | 6162 | 0.050 | 0.4992 | No | ||
| 19 | CCR8 | 19272 | 6509 | 0.043 | 0.4841 | No | ||
| 20 | GRPR | 24015 | 6711 | 0.039 | 0.4765 | No | ||
| 21 | MC2R | 23412 | 6941 | 0.035 | 0.4670 | No | ||
| 22 | CCR4 | 18978 | 7100 | 0.033 | 0.4612 | No | ||
| 23 | CCR6 | 1505 23390 | 7477 | 0.027 | 0.4431 | No | ||
| 24 | SSTR3 | 22218 | 7794 | 0.023 | 0.4279 | No | ||
| 25 | BDKRB2 | 21163 | 7844 | 0.022 | 0.4271 | No | ||
| 26 | NPY6R | 23567 | 7851 | 0.022 | 0.4286 | No | ||
| 27 | MC1R | 18428 | 8178 | 0.018 | 0.4124 | No | ||
| 28 | MC3R | 14721 | 8208 | 0.017 | 0.4123 | No | ||
| 29 | GALR3 | 9000 | 8335 | 0.016 | 0.4068 | No | ||
| 30 | TSHR | 5801 10223 | 8377 | 0.015 | 0.4058 | No | ||
| 31 | TACR2 | 20004 | 8378 | 0.015 | 0.4071 | No | ||
| 32 | MC4R | 13701 | 8447 | 0.015 | 0.4046 | No | ||
| 33 | GNRHR | 4791 | 8730 | 0.011 | 0.3903 | No | ||
| 34 | IL8RA | 13925 | 8869 | 0.010 | 0.3837 | No | ||
| 35 | NMBR | 9466 | 8931 | 0.009 | 0.3812 | No | ||
| 36 | PPYR1 | 21881 | 8990 | 0.009 | 0.3788 | No | ||
| 37 | BDKRB1 | 19418 | 9020 | 0.008 | 0.3779 | No | ||
| 38 | LHCGR | 9273 4994 | 9594 | 0.002 | 0.3472 | No | ||
| 39 | C3AR1 | 8668 | 9769 | 0.000 | 0.3378 | No | ||
| 40 | OPRK1 | 14300 | 9839 | -0.001 | 0.3341 | No | ||
| 41 | AVPR1B | 6441 | 10008 | -0.003 | 0.3253 | No | ||
| 42 | FSHR | 1492 22876 | 10042 | -0.003 | 0.3238 | No | ||
| 43 | NPY5R | 18866 | 10109 | -0.004 | 0.3205 | No | ||
| 44 | ATP8A1 | 4428 | 10111 | -0.004 | 0.3208 | No | ||
| 45 | NPY1R | 9482 | 10938 | -0.012 | 0.2772 | No | ||
| 46 | BRS3 | 24333 | 11113 | -0.014 | 0.2690 | No | ||
| 47 | EDNRA | 18834 | 11346 | -0.017 | 0.2579 | No | ||
| 48 | CCKBR | 8706 | 11511 | -0.019 | 0.2506 | No | ||
| 49 | FPRL1 | 8982 | 11722 | -0.022 | 0.2411 | No | ||
| 50 | GALR1 | 23397 | 11962 | -0.025 | 0.2302 | No | ||
| 51 | NPY2R | 343 | 12028 | -0.026 | 0.2288 | No | ||
| 52 | CCR10 | 20218 | 12052 | -0.026 | 0.2297 | No | ||
| 53 | AGTR2 | 24365 | 12124 | -0.027 | 0.2281 | No | ||
| 54 | GALR2 | 20587 | 12202 | -0.028 | 0.2262 | No | ||
| 55 | CCR3 | 19251 | 12305 | -0.029 | 0.2231 | No | ||
| 56 | SSTR1 | 21264 | 12559 | -0.033 | 0.2122 | No | ||
| 57 | CCR5 | 8754 4534 | 12625 | -0.034 | 0.2114 | No | ||
| 58 | EDNRB | 21728 3564 | 13624 | -0.054 | 0.1620 | No | ||
| 59 | AVPR1A | 19863 | 13627 | -0.054 | 0.1663 | No | ||
| 60 | CXCR6 | 19252 | 13908 | -0.061 | 0.1563 | No | ||
| 61 | MC5R | 23524 | 14239 | -0.072 | 0.1443 | No | ||
| 62 | CCR2 | 19250 | 14541 | -0.083 | 0.1349 | No | ||
| 63 | TACR3 | 15415 | 14683 | -0.090 | 0.1347 | No | ||
| 64 | OPRD1 | 15737 | 14767 | -0.094 | 0.1379 | No | ||
| 65 | CCR1 | 18951 | 15473 | -0.155 | 0.1127 | No | ||
| 66 | AVPR2 | 8646 | 15490 | -0.157 | 0.1246 | No | ||
| 67 | IL8RB | 8752 4533 | 15974 | -0.246 | 0.1187 | No | ||
| 68 | CXCR3 | 24093 | 16139 | -0.289 | 0.1335 | No |