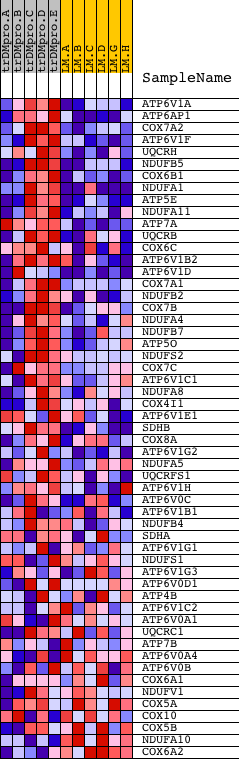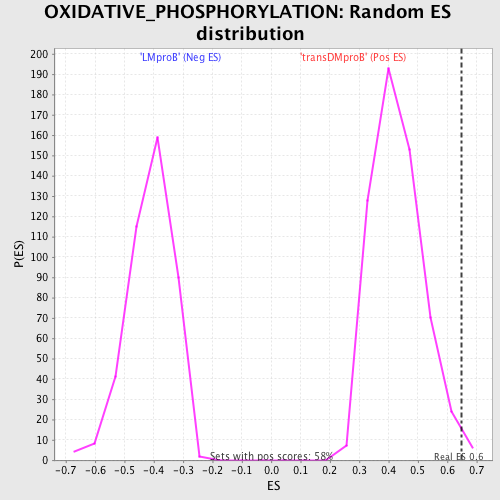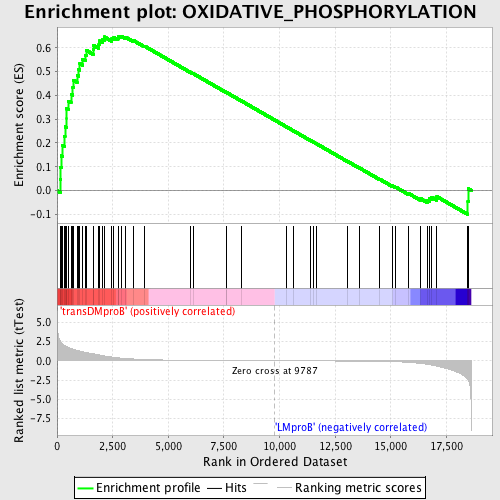
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | OXIDATIVE_PHOSPHORYLATION |
| Enrichment Score (ES) | 0.6491182 |
| Normalized Enrichment Score (NES) | 1.507506 |
| Nominal p-value | 0.0103270225 |
| FDR q-value | 0.37844434 |
| FWER p-Value | 0.969 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP6V1A | 8638 | 142 | 2.504 | 0.0466 | Yes | ||
| 2 | ATP6AP1 | 24296 | 162 | 2.422 | 0.0981 | Yes | ||
| 3 | COX7A2 | 19053 | 194 | 2.275 | 0.1457 | Yes | ||
| 4 | ATP6V1F | 12291 | 250 | 2.140 | 0.1891 | Yes | ||
| 5 | UQCRH | 12378 | 330 | 1.989 | 0.2280 | Yes | ||
| 6 | NDUFB5 | 12277 | 357 | 1.941 | 0.2686 | Yes | ||
| 7 | COX6B1 | 4283 8479 | 425 | 1.813 | 0.3043 | Yes | ||
| 8 | NDUFA1 | 24170 | 427 | 1.811 | 0.3435 | Yes | ||
| 9 | ATP5E | 14321 | 506 | 1.707 | 0.3763 | Yes | ||
| 10 | NDUFA11 | 12832 | 629 | 1.602 | 0.4044 | Yes | ||
| 11 | ATP7A | 4425 | 707 | 1.526 | 0.4334 | Yes | ||
| 12 | UQCRB | 12545 | 735 | 1.494 | 0.4643 | Yes | ||
| 13 | COX6C | 8775 | 900 | 1.344 | 0.4846 | Yes | ||
| 14 | ATP6V1B2 | 18599 | 973 | 1.288 | 0.5086 | Yes | ||
| 15 | ATP6V1D | 7872 | 1021 | 1.251 | 0.5332 | Yes | ||
| 16 | COX7A1 | 4554 | 1141 | 1.175 | 0.5522 | Yes | ||
| 17 | NDUFB2 | 7542 | 1281 | 1.081 | 0.5682 | Yes | ||
| 18 | COX7B | 7260 | 1326 | 1.043 | 0.5884 | Yes | ||
| 19 | NDUFA4 | 9451 | 1624 | 0.903 | 0.5920 | Yes | ||
| 20 | NDUFB7 | 12429 | 1640 | 0.896 | 0.6106 | Yes | ||
| 21 | ATP5O | 22539 | 1878 | 0.758 | 0.6142 | Yes | ||
| 22 | NDUFS2 | 4089 13760 | 1905 | 0.742 | 0.6289 | Yes | ||
| 23 | COX7C | 8776 | 2035 | 0.672 | 0.6365 | Yes | ||
| 24 | ATP6V1C1 | 22487 12329 | 2121 | 0.634 | 0.6457 | Yes | ||
| 25 | NDUFA8 | 14605 | 2462 | 0.483 | 0.6378 | Yes | ||
| 26 | COX4I1 | 18444 | 2550 | 0.449 | 0.6429 | Yes | ||
| 27 | ATP6V1E1 | 4423 | 2737 | 0.382 | 0.6411 | Yes | ||
| 28 | SDHB | 2348 12566 14880 | 2767 | 0.373 | 0.6476 | Yes | ||
| 29 | COX8A | 8777 | 2879 | 0.344 | 0.6491 | Yes | ||
| 30 | ATP6V1G2 | 23257 1534 | 3086 | 0.289 | 0.6443 | No | ||
| 31 | NDUFA5 | 1076 7544 | 3441 | 0.223 | 0.6300 | No | ||
| 32 | UQCRFS1 | 21499 | 3927 | 0.158 | 0.6073 | No | ||
| 33 | ATP6V1H | 14301 | 5973 | 0.055 | 0.4984 | No | ||
| 34 | ATP6V0C | 8643 | 6122 | 0.051 | 0.4915 | No | ||
| 35 | ATP6V1B1 | 8499 | 7591 | 0.025 | 0.4130 | No | ||
| 36 | NDUFB4 | 22596 7541 | 7626 | 0.025 | 0.4117 | No | ||
| 37 | SDHA | 12436 | 8271 | 0.017 | 0.3773 | No | ||
| 38 | ATP6V1G1 | 12322 | 10294 | -0.006 | 0.2685 | No | ||
| 39 | NDUFS1 | 13940 | 10640 | -0.009 | 0.2501 | No | ||
| 40 | ATP6V1G3 | 14107 | 11399 | -0.018 | 0.2097 | No | ||
| 41 | ATP6V0D1 | 3895 8639 3920 | 11540 | -0.019 | 0.2026 | No | ||
| 42 | ATP4B | 18928 | 11640 | -0.021 | 0.1977 | No | ||
| 43 | ATP6V1C2 | 21098 | 13050 | -0.042 | 0.1227 | No | ||
| 44 | ATP6V0A1 | 8640 4424 1197 | 13586 | -0.053 | 0.0950 | No | ||
| 45 | UQCRC1 | 10259 | 14490 | -0.081 | 0.0481 | No | ||
| 46 | ATP7B | 18918 4426 4427 8641 | 15065 | -0.116 | 0.0197 | No | ||
| 47 | ATP6V0A4 | 8934 | 15216 | -0.128 | 0.0144 | No | ||
| 48 | ATP6V0B | 15786 | 15794 | -0.208 | -0.0122 | No | ||
| 49 | COX6A1 | 8774 4553 | 16325 | -0.342 | -0.0333 | No | ||
| 50 | NDUFV1 | 23953 | 16637 | -0.452 | -0.0403 | No | ||
| 51 | COX5A | 19431 | 16734 | -0.506 | -0.0345 | No | ||
| 52 | COX10 | 20408 | 16825 | -0.545 | -0.0276 | No | ||
| 53 | COX5B | 4552 | 17069 | -0.693 | -0.0256 | No | ||
| 54 | NDUFA10 | 7420 | 18465 | -2.476 | -0.0471 | No | ||
| 55 | COX6A2 | 17605 | 18485 | -2.548 | 0.0071 | No |