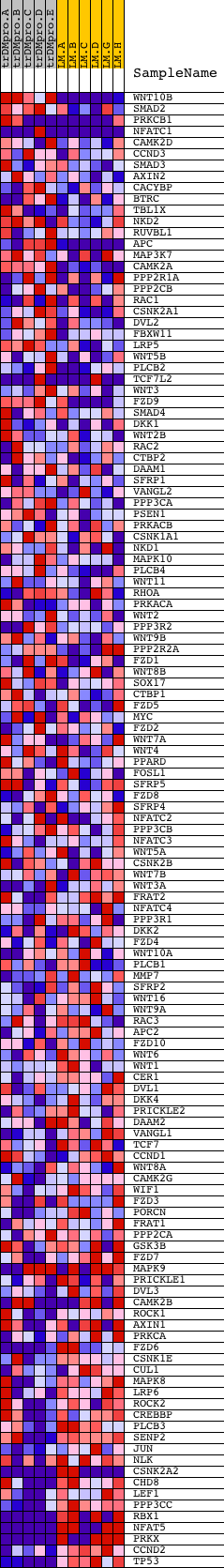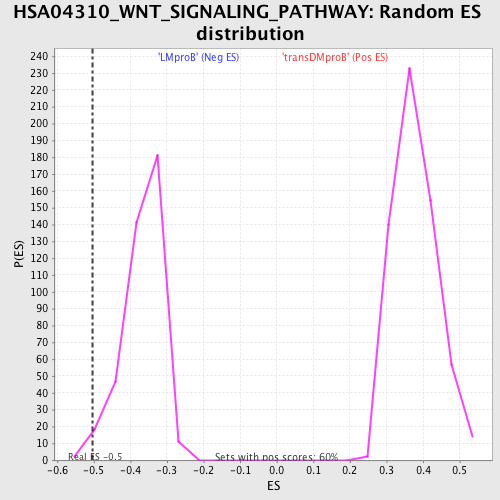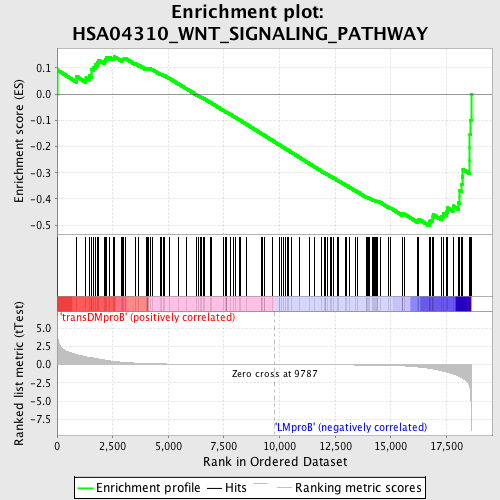
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | HSA04310_WNT_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.5031088 |
| Normalized Enrichment Score (NES) | -1.3743987 |
| Nominal p-value | 0.0175 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | WNT10B | 5874 | 2 | 5.814 | 0.0927 | No | ||
| 2 | SMAD2 | 23511 | 864 | 1.376 | 0.0681 | No | ||
| 3 | PRKCB1 | 1693 9574 | 1297 | 1.068 | 0.0618 | No | ||
| 4 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 1446 | 0.991 | 0.0696 | No | ||
| 5 | CAMK2D | 4232 | 1556 | 0.941 | 0.0788 | No | ||
| 6 | CCND3 | 4489 4490 | 1557 | 0.941 | 0.0938 | No | ||
| 7 | SMAD3 | 19084 | 1649 | 0.891 | 0.1031 | No | ||
| 8 | AXIN2 | 20618 | 1729 | 0.842 | 0.1123 | No | ||
| 9 | CACYBP | 4468 | 1824 | 0.788 | 0.1198 | No | ||
| 10 | BTRC | 4459 | 1873 | 0.760 | 0.1293 | No | ||
| 11 | TBL1X | 5628 | 2137 | 0.623 | 0.1250 | No | ||
| 12 | NKD2 | 7798 | 2167 | 0.605 | 0.1331 | No | ||
| 13 | RUVBL1 | 7131 7130 | 2221 | 0.574 | 0.1394 | No | ||
| 14 | APC | 4396 2022 | 2338 | 0.526 | 0.1415 | No | ||
| 15 | MAP3K7 | 16255 | 2546 | 0.451 | 0.1375 | No | ||
| 16 | CAMK2A | 2024 23541 1980 | 2586 | 0.432 | 0.1423 | No | ||
| 17 | PPP2R1A | 11951 | 2908 | 0.337 | 0.1303 | No | ||
| 18 | PPP2CB | 18636 | 2947 | 0.323 | 0.1334 | No | ||
| 19 | RAC1 | 16302 | 3005 | 0.309 | 0.1353 | No | ||
| 20 | CSNK2A1 | 14797 | 3073 | 0.293 | 0.1364 | No | ||
| 21 | DVL2 | 20813 | 3517 | 0.211 | 0.1158 | No | ||
| 22 | FBXW11 | 20926 | 3676 | 0.191 | 0.1103 | No | ||
| 23 | LRP5 | 23948 9285 | 4008 | 0.151 | 0.0948 | No | ||
| 24 | WNT5B | 17022 | 4040 | 0.148 | 0.0954 | No | ||
| 25 | PLCB2 | 5262 | 4054 | 0.147 | 0.0971 | No | ||
| 26 | TCF7L2 | 10048 5646 | 4074 | 0.145 | 0.0984 | No | ||
| 27 | WNT3 | 20635 | 4109 | 0.141 | 0.0988 | No | ||
| 28 | FZD9 | 16345 | 4189 | 0.134 | 0.0966 | No | ||
| 29 | SMAD4 | 5058 | 4275 | 0.126 | 0.0940 | No | ||
| 30 | DKK1 | 23700 | 4659 | 0.103 | 0.0750 | No | ||
| 31 | WNT2B | 15214 1838 | 4681 | 0.101 | 0.0754 | No | ||
| 32 | RAC2 | 22217 | 4775 | 0.096 | 0.0719 | No | ||
| 33 | CTBP2 | 17591 1137 | 4842 | 0.092 | 0.0698 | No | ||
| 34 | DAAM1 | 5536 | 5040 | 0.084 | 0.0605 | No | ||
| 35 | SFRP1 | 9805 | 5437 | 0.070 | 0.0402 | No | ||
| 36 | VANGL2 | 4109 8220 | 5796 | 0.060 | 0.0218 | No | ||
| 37 | PPP3CA | 1863 5284 | 6257 | 0.048 | -0.0023 | No | ||
| 38 | PSEN1 | 5297 2125 9630 | 6334 | 0.046 | -0.0057 | No | ||
| 39 | PRKACB | 15140 | 6459 | 0.044 | -0.0117 | No | ||
| 40 | CSNK1A1 | 8204 | 6501 | 0.043 | -0.0132 | No | ||
| 41 | NKD1 | 8228 | 6597 | 0.041 | -0.0177 | No | ||
| 42 | MAPK10 | 11169 | 6603 | 0.041 | -0.0173 | No | ||
| 43 | PLCB4 | 9586 | 6893 | 0.036 | -0.0324 | No | ||
| 44 | WNT11 | 5876 | 6929 | 0.035 | -0.0337 | No | ||
| 45 | RHOA | 8624 4409 4410 | 7462 | 0.027 | -0.0620 | No | ||
| 46 | PRKACA | 18549 3844 | 7558 | 0.026 | -0.0668 | No | ||
| 47 | WNT2 | 17212 | 7603 | 0.025 | -0.0687 | No | ||
| 48 | PPP3R2 | 9612 | 7627 | 0.025 | -0.0696 | No | ||
| 49 | WNT9B | 20193 | 7632 | 0.025 | -0.0694 | No | ||
| 50 | PPP2R2A | 3222 21774 | 7792 | 0.023 | -0.0776 | No | ||
| 51 | FZD1 | 16923 | 7931 | 0.021 | -0.0848 | No | ||
| 52 | WNT8B | 23841 | 8034 | 0.019 | -0.0900 | No | ||
| 53 | SOX17 | 14005 | 8214 | 0.017 | -0.0994 | No | ||
| 54 | CTBP1 | 16566 | 8222 | 0.017 | -0.0995 | No | ||
| 55 | FZD5 | 4743 | 8505 | 0.014 | -0.1145 | No | ||
| 56 | MYC | 22465 9435 | 9165 | 0.007 | -0.1501 | No | ||
| 57 | FZD2 | 12179 | 9188 | 0.006 | -0.1512 | No | ||
| 58 | WNT7A | 17068 | 9214 | 0.006 | -0.1524 | No | ||
| 59 | WNT4 | 16025 | 9305 | 0.005 | -0.1572 | No | ||
| 60 | PPARD | 9606 | 9323 | 0.005 | -0.1580 | No | ||
| 61 | FOSL1 | 23779 | 9674 | 0.001 | -0.1770 | No | ||
| 62 | SFRP5 | 23674 | 10002 | -0.003 | -0.1946 | No | ||
| 63 | FZD8 | 8990 | 10004 | -0.003 | -0.1946 | No | ||
| 64 | SFRP4 | 9806 | 10083 | -0.003 | -0.1988 | No | ||
| 65 | NFATC2 | 5168 2866 | 10190 | -0.005 | -0.2044 | No | ||
| 66 | PPP3CB | 5285 | 10275 | -0.005 | -0.2089 | No | ||
| 67 | NFATC3 | 5169 | 10340 | -0.006 | -0.2123 | No | ||
| 68 | WNT5A | 22066 5880 | 10414 | -0.007 | -0.2161 | No | ||
| 69 | CSNK2B | 23008 | 10542 | -0.008 | -0.2228 | No | ||
| 70 | WNT7B | 22172 | 10872 | -0.012 | -0.2405 | No | ||
| 71 | WNT3A | 20431 | 11325 | -0.017 | -0.2646 | No | ||
| 72 | FRAT2 | 23677 | 11549 | -0.019 | -0.2764 | No | ||
| 73 | NFATC4 | 22002 | 11905 | -0.024 | -0.2952 | No | ||
| 74 | PPP3R1 | 9611 489 | 12037 | -0.026 | -0.3019 | No | ||
| 75 | DKK2 | 15418 | 12042 | -0.026 | -0.3017 | No | ||
| 76 | FZD4 | 18193 | 12081 | -0.026 | -0.3033 | No | ||
| 77 | WNT10A | 14221 | 12134 | -0.027 | -0.3057 | No | ||
| 78 | PLCB1 | 14832 2821 | 12286 | -0.029 | -0.3134 | No | ||
| 79 | MMP7 | 19568 | 12331 | -0.030 | -0.3153 | No | ||
| 80 | SFRP2 | 15564 | 12400 | -0.031 | -0.3185 | No | ||
| 81 | WNT16 | 17512 1152 | 12580 | -0.033 | -0.3277 | No | ||
| 82 | WNT9A | 5709 | 12649 | -0.035 | -0.3308 | No | ||
| 83 | RAC3 | 20561 | 12977 | -0.040 | -0.3478 | No | ||
| 84 | APC2 | 10704 | 13023 | -0.041 | -0.3496 | No | ||
| 85 | FZD10 | 13565 16693 | 13141 | -0.043 | -0.3553 | No | ||
| 86 | WNT6 | 5881 | 13403 | -0.048 | -0.3686 | No | ||
| 87 | WNT1 | 22371 | 13503 | -0.051 | -0.3731 | No | ||
| 88 | CER1 | 15853 | 13887 | -0.061 | -0.3929 | No | ||
| 89 | DVL1 | 2412 15960 | 13925 | -0.062 | -0.3939 | No | ||
| 90 | DKK4 | 18653 | 13929 | -0.062 | -0.3931 | No | ||
| 91 | PRICKLE2 | 10837 | 13975 | -0.063 | -0.3945 | No | ||
| 92 | DAAM2 | 22945 176 | 14036 | -0.065 | -0.3967 | No | ||
| 93 | VANGL1 | 6034 | 14186 | -0.070 | -0.4037 | No | ||
| 94 | TCF7 | 1467 20466 | 14232 | -0.071 | -0.4049 | No | ||
| 95 | CCND1 | 4487 4488 8707 17535 | 14255 | -0.072 | -0.4050 | No | ||
| 96 | WNT8A | 1978 23603 | 14332 | -0.075 | -0.4079 | No | ||
| 97 | CAMK2G | 21905 | 14362 | -0.076 | -0.4083 | No | ||
| 98 | WIF1 | 19866 | 14406 | -0.078 | -0.4093 | No | ||
| 99 | FZD3 | 21784 | 14422 | -0.079 | -0.4089 | No | ||
| 100 | PORCN | 24190 | 14522 | -0.082 | -0.4129 | No | ||
| 101 | FRAT1 | 3681 23856 | 14885 | -0.102 | -0.4309 | No | ||
| 102 | PPP2CA | 20890 | 15002 | -0.111 | -0.4354 | No | ||
| 103 | GSK3B | 22761 | 15502 | -0.158 | -0.4599 | No | ||
| 104 | FZD7 | 14242 | 15503 | -0.158 | -0.4573 | No | ||
| 105 | MAPK9 | 1233 20903 1383 | 15505 | -0.159 | -0.4549 | No | ||
| 106 | PRICKLE1 | 8379 | 15622 | -0.176 | -0.4583 | No | ||
| 107 | DVL3 | 8873 22821 | 16191 | -0.303 | -0.4842 | No | ||
| 108 | CAMK2B | 20536 | 16235 | -0.314 | -0.4815 | No | ||
| 109 | ROCK1 | 5386 | 16262 | -0.322 | -0.4778 | No | ||
| 110 | AXIN1 | 1579 23330 | 16731 | -0.504 | -0.4951 | Yes | ||
| 111 | PRKCA | 20174 | 16737 | -0.507 | -0.4872 | Yes | ||
| 112 | FZD6 | 22486 | 16773 | -0.524 | -0.4808 | Yes | ||
| 113 | CSNK1E | 6570 2211 11332 | 16889 | -0.582 | -0.4777 | Yes | ||
| 114 | CUL1 | 17462 | 16891 | -0.584 | -0.4685 | Yes | ||
| 115 | MAPK8 | 6459 | 16914 | -0.593 | -0.4602 | Yes | ||
| 116 | LRP6 | 9286 | 17270 | -0.837 | -0.4660 | Yes | ||
| 117 | ROCK2 | 21309 5387 | 17355 | -0.908 | -0.4560 | Yes | ||
| 118 | CREBBP | 22682 8783 | 17516 | -1.011 | -0.4486 | Yes | ||
| 119 | PLCB3 | 23799 | 17555 | -1.037 | -0.4341 | Yes | ||
| 120 | SENP2 | 7990 | 17798 | -1.280 | -0.4267 | Yes | ||
| 121 | JUN | 15832 | 18032 | -1.559 | -0.4144 | Yes | ||
| 122 | NLK | 5179 5178 | 18082 | -1.602 | -0.3915 | Yes | ||
| 123 | CSNK2A2 | 4567 8808 | 18083 | -1.602 | -0.3659 | Yes | ||
| 124 | CHD8 | 12581 | 18189 | -1.768 | -0.3434 | Yes | ||
| 125 | LEF1 | 1860 15420 | 18218 | -1.821 | -0.3158 | Yes | ||
| 126 | PPP3CC | 21763 | 18241 | -1.882 | -0.2870 | Yes | ||
| 127 | RBX1 | 12130 7123 | 18550 | -3.114 | -0.2539 | Yes | ||
| 128 | NFAT5 | 3921 7037 12036 | 18552 | -3.129 | -0.2040 | Yes | ||
| 129 | PRKX | 9619 5290 | 18555 | -3.145 | -0.1539 | Yes | ||
| 130 | CCND2 | 16987 | 18573 | -3.414 | -0.1003 | Yes | ||
| 131 | TP53 | 20822 | 18610 | -6.426 | 0.0003 | Yes |