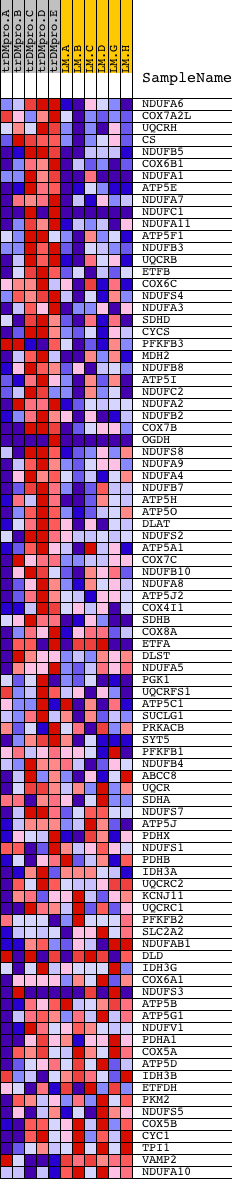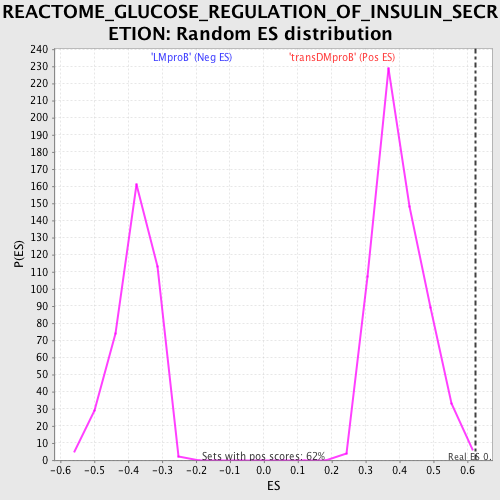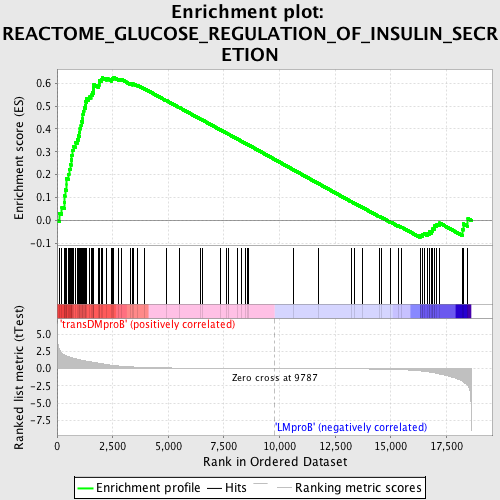
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | REACTOME_GLUCOSE_REGULATION_OF_INSULIN_SECRETION |
| Enrichment Score (ES) | 0.6250255 |
| Normalized Enrichment Score (NES) | 1.5564576 |
| Nominal p-value | 0.0048701297 |
| FDR q-value | 0.6060722 |
| FWER p-Value | 0.909 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NDUFA6 | 12473 | 107 | 2.667 | 0.0321 | Yes | ||
| 2 | COX7A2L | 9819 | 214 | 2.226 | 0.0579 | Yes | ||
| 3 | UQCRH | 12378 | 330 | 1.989 | 0.0800 | Yes | ||
| 4 | CS | 19839 | 345 | 1.955 | 0.1070 | Yes | ||
| 5 | NDUFB5 | 12277 | 357 | 1.941 | 0.1339 | Yes | ||
| 6 | COX6B1 | 4283 8479 | 425 | 1.813 | 0.1560 | Yes | ||
| 7 | NDUFA1 | 24170 | 427 | 1.811 | 0.1817 | Yes | ||
| 8 | ATP5E | 14321 | 506 | 1.707 | 0.2017 | Yes | ||
| 9 | NDUFA7 | 12347 | 537 | 1.673 | 0.2238 | Yes | ||
| 10 | NDUFC1 | 7287 12339 | 594 | 1.631 | 0.2439 | Yes | ||
| 11 | NDUFA11 | 12832 | 629 | 1.602 | 0.2648 | Yes | ||
| 12 | ATP5F1 | 15212 | 656 | 1.576 | 0.2858 | Yes | ||
| 13 | NDUFB3 | 12362 | 677 | 1.551 | 0.3067 | Yes | ||
| 14 | UQCRB | 12545 | 735 | 1.494 | 0.3249 | Yes | ||
| 15 | ETFB | 18279 | 837 | 1.400 | 0.3393 | Yes | ||
| 16 | COX6C | 8775 | 900 | 1.344 | 0.3550 | Yes | ||
| 17 | NDUFS4 | 9452 5157 3173 | 969 | 1.290 | 0.3697 | Yes | ||
| 18 | NDUFA3 | 18410 | 1012 | 1.256 | 0.3852 | Yes | ||
| 19 | SDHD | 19125 | 1026 | 1.248 | 0.4022 | Yes | ||
| 20 | CYCS | 8821 | 1071 | 1.216 | 0.4171 | Yes | ||
| 21 | PFKFB3 | 2748 | 1091 | 1.204 | 0.4332 | Yes | ||
| 22 | MDH2 | 9399 | 1152 | 1.168 | 0.4465 | Yes | ||
| 23 | NDUFB8 | 7418 | 1159 | 1.161 | 0.4627 | Yes | ||
| 24 | ATP5I | 8637 | 1170 | 1.149 | 0.4784 | Yes | ||
| 25 | NDUFC2 | 18183 | 1209 | 1.127 | 0.4924 | Yes | ||
| 26 | NDUFA2 | 9450 | 1265 | 1.089 | 0.5049 | Yes | ||
| 27 | NDUFB2 | 7542 | 1281 | 1.081 | 0.5194 | Yes | ||
| 28 | COX7B | 7260 | 1326 | 1.043 | 0.5319 | Yes | ||
| 29 | OGDH | 9502 1412 | 1433 | 0.996 | 0.5403 | Yes | ||
| 30 | NDUFS8 | 23950 | 1524 | 0.958 | 0.5490 | Yes | ||
| 31 | NDUFA9 | 12288 | 1595 | 0.923 | 0.5583 | Yes | ||
| 32 | NDUFA4 | 9451 | 1624 | 0.903 | 0.5696 | Yes | ||
| 33 | NDUFB7 | 12429 | 1640 | 0.896 | 0.5815 | Yes | ||
| 34 | ATP5H | 12948 | 1651 | 0.890 | 0.5936 | Yes | ||
| 35 | ATP5O | 22539 | 1878 | 0.758 | 0.5922 | Yes | ||
| 36 | DLAT | 19123 | 1886 | 0.753 | 0.6025 | Yes | ||
| 37 | NDUFS2 | 4089 13760 | 1905 | 0.742 | 0.6120 | Yes | ||
| 38 | ATP5A1 | 23505 | 1993 | 0.692 | 0.6172 | Yes | ||
| 39 | COX7C | 8776 | 2035 | 0.672 | 0.6245 | Yes | ||
| 40 | NDUFB10 | 23084 | 2227 | 0.572 | 0.6223 | Yes | ||
| 41 | NDUFA8 | 14605 | 2462 | 0.483 | 0.6165 | Yes | ||
| 42 | ATP5J2 | 12186 | 2483 | 0.476 | 0.6222 | Yes | ||
| 43 | COX4I1 | 18444 | 2550 | 0.449 | 0.6250 | Yes | ||
| 44 | SDHB | 2348 12566 14880 | 2767 | 0.373 | 0.6187 | No | ||
| 45 | COX8A | 8777 | 2879 | 0.344 | 0.6175 | No | ||
| 46 | ETFA | 4292 8495 3155 | 3312 | 0.244 | 0.5977 | No | ||
| 47 | DLST | 8131 | 3410 | 0.228 | 0.5957 | No | ||
| 48 | NDUFA5 | 1076 7544 | 3441 | 0.223 | 0.5972 | No | ||
| 49 | PGK1 | 9557 5244 | 3613 | 0.197 | 0.5908 | No | ||
| 50 | UQCRFS1 | 21499 | 3927 | 0.158 | 0.5762 | No | ||
| 51 | ATP5C1 | 8635 | 4914 | 0.089 | 0.5242 | No | ||
| 52 | SUCLG1 | 17409 1059 | 5507 | 0.067 | 0.4932 | No | ||
| 53 | PRKACB | 15140 | 6459 | 0.044 | 0.4425 | No | ||
| 54 | SYT5 | 17986 | 6531 | 0.043 | 0.4393 | No | ||
| 55 | PFKFB1 | 9552 | 7345 | 0.029 | 0.3958 | No | ||
| 56 | NDUFB4 | 22596 7541 | 7626 | 0.025 | 0.3810 | No | ||
| 57 | ABCC8 | 9941 11649 | 7711 | 0.024 | 0.3768 | No | ||
| 58 | UQCR | 12382 | 8090 | 0.018 | 0.3567 | No | ||
| 59 | SDHA | 12436 | 8271 | 0.017 | 0.3472 | No | ||
| 60 | NDUFS7 | 19946 | 8476 | 0.014 | 0.3364 | No | ||
| 61 | ATP5J | 880 22555 | 8543 | 0.014 | 0.3330 | No | ||
| 62 | PDHX | 14510 | 8608 | 0.013 | 0.3298 | No | ||
| 63 | NDUFS1 | 13940 | 10640 | -0.009 | 0.2203 | No | ||
| 64 | PDHB | 12670 7548 | 11762 | -0.022 | 0.1601 | No | ||
| 65 | IDH3A | 19447 | 13210 | -0.045 | 0.0826 | No | ||
| 66 | UQCRC2 | 18102 | 13386 | -0.048 | 0.0739 | No | ||
| 67 | KCNJ11 | 17822 | 13734 | -0.057 | 0.0559 | No | ||
| 68 | UQCRC1 | 10259 | 14490 | -0.081 | 0.0163 | No | ||
| 69 | PFKFB2 | 4116 5242 | 14574 | -0.084 | 0.0131 | No | ||
| 70 | SLC2A2 | 15623 1880 1855 | 14983 | -0.109 | -0.0074 | No | ||
| 71 | NDUFAB1 | 7667 | 15329 | -0.139 | -0.0241 | No | ||
| 72 | DLD | 2097 21090 | 15499 | -0.158 | -0.0310 | No | ||
| 73 | IDH3G | 24132 | 16316 | -0.338 | -0.0702 | No | ||
| 74 | COX6A1 | 8774 4553 | 16325 | -0.342 | -0.0658 | No | ||
| 75 | NDUFS3 | 7553 12683 | 16422 | -0.378 | -0.0656 | No | ||
| 76 | ATP5B | 19846 | 16431 | -0.383 | -0.0606 | No | ||
| 77 | ATP5G1 | 8636 | 16499 | -0.407 | -0.0584 | No | ||
| 78 | NDUFV1 | 23953 | 16637 | -0.452 | -0.0594 | No | ||
| 79 | PDHA1 | 24020 | 16724 | -0.500 | -0.0570 | No | ||
| 80 | COX5A | 19431 | 16734 | -0.506 | -0.0503 | No | ||
| 81 | ATP5D | 19949 | 16843 | -0.556 | -0.0482 | No | ||
| 82 | IDH3B | 9299 | 16859 | -0.566 | -0.0410 | No | ||
| 83 | ETFDH | 15313 | 16890 | -0.583 | -0.0343 | No | ||
| 84 | PKM2 | 3642 9573 | 16970 | -0.629 | -0.0297 | No | ||
| 85 | NDUFS5 | 9296 | 16976 | -0.630 | -0.0210 | No | ||
| 86 | COX5B | 4552 | 17069 | -0.693 | -0.0161 | No | ||
| 87 | CYC1 | 12348 | 17188 | -0.772 | -0.0115 | No | ||
| 88 | TPI1 | 5795 10212 | 18230 | -1.859 | -0.0413 | No | ||
| 89 | VAMP2 | 20826 | 18251 | -1.897 | -0.0155 | No | ||
| 90 | NDUFA10 | 7420 | 18465 | -2.476 | 0.0082 | No |