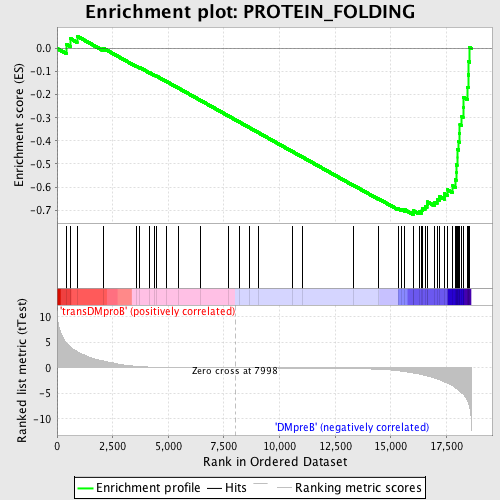
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | PROTEIN_FOLDING |
| Enrichment Score (ES) | -0.71884567 |
| Normalized Enrichment Score (NES) | -1.6488146 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.022333674 |
| FWER p-Value | 0.347 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | LRPAP1 | 5005 | 411 | 4.921 | 0.0190 | No | ||
| 2 | AIP | 23955 | 602 | 4.063 | 0.0427 | No | ||
| 3 | TBCC | 23215 | 897 | 3.189 | 0.0535 | No | ||
| 4 | TBCA | 10037 | 2075 | 1.334 | 0.0013 | No | ||
| 5 | PDIA6 | 21308 | 3579 | 0.290 | -0.0773 | No | ||
| 6 | HSPA4L | 5214 | 3719 | 0.252 | -0.0826 | No | ||
| 7 | FKBP5 | 23051 | 4174 | 0.151 | -0.1058 | No | ||
| 8 | PPIA | 1284 11188 | 4376 | 0.122 | -0.1156 | No | ||
| 9 | CCT6B | 20326 | 4457 | 0.113 | -0.1190 | No | ||
| 10 | PDIA2 | 23062 22604 1702 | 4913 | 0.073 | -0.1429 | No | ||
| 11 | DNAJA1 | 4878 16242 2345 | 5445 | 0.048 | -0.1711 | No | ||
| 12 | DNAJB6 | 6246 3538 | 6459 | 0.023 | -0.2255 | No | ||
| 13 | MKKS | 2946 14416 | 7719 | 0.004 | -0.2933 | No | ||
| 14 | APCS | 13748 | 8184 | -0.003 | -0.3182 | No | ||
| 15 | FKBP9 | 17433 | 8648 | -0.009 | -0.3431 | No | ||
| 16 | NFYC | 9460 | 9047 | -0.014 | -0.3644 | No | ||
| 17 | FKBP6 | 16344 | 10586 | -0.038 | -0.4469 | No | ||
| 18 | TBCE | 21543 | 11035 | -0.047 | -0.4707 | No | ||
| 19 | ERO1L | 6930 11924 6929 | 13327 | -0.141 | -0.5929 | No | ||
| 20 | PIGK | 1815 6834 11629 | 14435 | -0.271 | -0.6503 | No | ||
| 21 | APOA2 | 8615 4044 | 15323 | -0.544 | -0.6935 | No | ||
| 22 | STUB1 | 23070 | 15472 | -0.640 | -0.6961 | No | ||
| 23 | UGCGL1 | 6721 | 15631 | -0.730 | -0.6985 | No | ||
| 24 | BAG5 | 20981 192 | 16009 | -1.003 | -0.7105 | Yes | ||
| 25 | DNAJA3 | 1732 13518 | 16014 | -1.005 | -0.7023 | Yes | ||
| 26 | PIN4 | 12817 | 16277 | -1.204 | -0.7063 | Yes | ||
| 27 | CLPX | 19403 | 16400 | -1.335 | -0.7017 | Yes | ||
| 28 | TXNDC4 | 15886 | 16424 | -1.354 | -0.6917 | Yes | ||
| 29 | LMAN1 | 12867 | 16539 | -1.502 | -0.6853 | Yes | ||
| 30 | CLN3 | 17635 | 16628 | -1.574 | -0.6768 | Yes | ||
| 31 | HSP90AA1 | 4883 4882 9131 | 16644 | -1.583 | -0.6644 | Yes | ||
| 32 | CCT4 | 8710 | 16964 | -1.978 | -0.6651 | Yes | ||
| 33 | TBCD | 20555 | 17079 | -2.148 | -0.6533 | Yes | ||
| 34 | TOR1A | 11395 2909 | 17180 | -2.263 | -0.6397 | Yes | ||
| 35 | CCT7 | 17385 | 17414 | -2.723 | -0.6295 | Yes | ||
| 36 | ARL2 | 23992 | 17551 | -2.992 | -0.6119 | Yes | ||
| 37 | DNAJC7 | 20227 | 17784 | -3.518 | -0.5950 | Yes | ||
| 38 | AHSA1 | 21192 | 17899 | -3.870 | -0.5687 | Yes | ||
| 39 | TTC1 | 7342 | 17935 | -3.997 | -0.5372 | Yes | ||
| 40 | BAG4 | 7431 | 17942 | -4.019 | -0.5040 | Yes | ||
| 41 | ST13 | 12865 | 17978 | -4.133 | -0.4713 | Yes | ||
| 42 | PPIH | 12287 | 17979 | -4.134 | -0.4367 | Yes | ||
| 43 | FKBP4 | 4726 | 18042 | -4.347 | -0.4038 | Yes | ||
| 44 | SEP15 | 13527 1895 15398 | 18082 | -4.516 | -0.3681 | Yes | ||
| 45 | BAG2 | 13985 | 18106 | -4.605 | -0.3309 | Yes | ||
| 46 | PFDN4 | 4260 | 18191 | -4.937 | -0.2941 | Yes | ||
| 47 | GLRX2 | 12795 | 18249 | -5.094 | -0.2546 | Yes | ||
| 48 | ERP29 | 12524 | 18258 | -5.122 | -0.2122 | Yes | ||
| 49 | RUVBL2 | 9766 | 18447 | -6.380 | -0.1690 | Yes | ||
| 50 | CCT6A | 16689 | 18478 | -6.752 | -0.1142 | Yes | ||
| 51 | HSPE1 | 9133 | 18500 | -6.948 | -0.0573 | Yes | ||
| 52 | CCT3 | 8709 | 18546 | -7.596 | 0.0038 | Yes |

