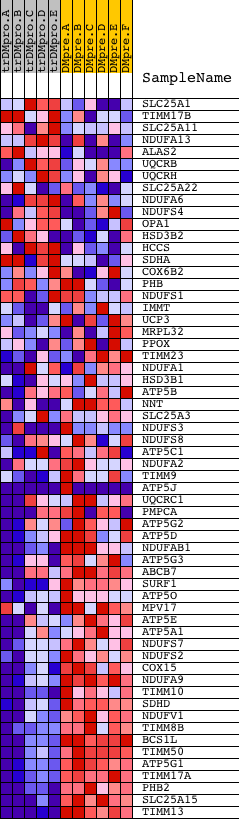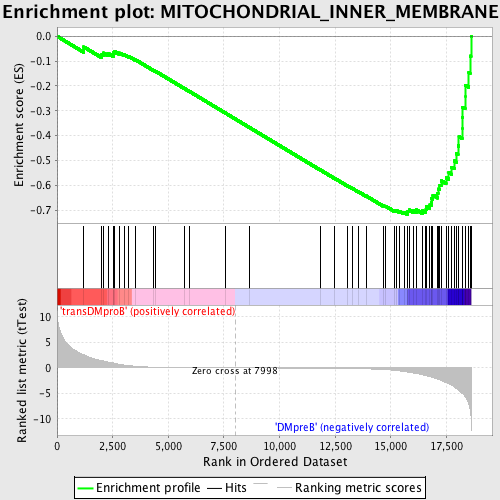
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | MITOCHONDRIAL_INNER_MEMBRANE |
| Enrichment Score (ES) | -0.7178924 |
| Normalized Enrichment Score (NES) | -1.6649857 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.018457137 |
| FWER p-Value | 0.26 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SLC25A1 | 22645 | 1200 | 2.530 | -0.0431 | No | ||
| 2 | TIMM17B | 24390 | 2010 | 1.393 | -0.0748 | No | ||
| 3 | SLC25A11 | 20366 1368 | 2082 | 1.328 | -0.0673 | No | ||
| 4 | NDUFA13 | 7400 | 2310 | 1.108 | -0.0700 | No | ||
| 5 | ALAS2 | 24229 | 2526 | 0.943 | -0.0736 | No | ||
| 6 | UQCRB | 12545 | 2534 | 0.936 | -0.0660 | No | ||
| 7 | UQCRH | 12378 | 2594 | 0.878 | -0.0616 | No | ||
| 8 | SLC25A22 | 12671 | 2809 | 0.684 | -0.0673 | No | ||
| 9 | NDUFA6 | 12473 | 3007 | 0.551 | -0.0732 | No | ||
| 10 | NDUFS4 | 9452 5157 3173 | 3225 | 0.439 | -0.0812 | No | ||
| 11 | OPA1 | 13169 22800 7894 | 3533 | 0.307 | -0.0951 | No | ||
| 12 | HSD3B2 | 4873 4874 15229 4875 9126 | 4323 | 0.132 | -0.1365 | No | ||
| 13 | HCCS | 9079 4842 | 4443 | 0.114 | -0.1419 | No | ||
| 14 | SDHA | 12436 | 5741 | 0.039 | -0.2115 | No | ||
| 15 | COX6B2 | 17983 4110 | 5948 | 0.034 | -0.2223 | No | ||
| 16 | PHB | 9558 | 7582 | 0.005 | -0.3103 | No | ||
| 17 | NDUFS1 | 13940 | 8627 | -0.008 | -0.3664 | No | ||
| 18 | IMMT | 367 8029 | 11818 | -0.066 | -0.5378 | No | ||
| 19 | UCP3 | 3972 18175 | 12478 | -0.092 | -0.5725 | No | ||
| 20 | MRPL32 | 7961 | 13055 | -0.122 | -0.6025 | No | ||
| 21 | PPOX | 13761 4104 | 13283 | -0.138 | -0.6136 | No | ||
| 22 | TIMM23 | 7004 5768 | 13546 | -0.160 | -0.6263 | No | ||
| 23 | NDUFA1 | 24170 | 13888 | -0.194 | -0.6430 | No | ||
| 24 | HSD3B1 | 15227 | 14654 | -0.318 | -0.6815 | No | ||
| 25 | ATP5B | 19846 | 14751 | -0.346 | -0.6838 | No | ||
| 26 | NNT | 3273 5181 9471 3238 | 15173 | -0.484 | -0.7023 | No | ||
| 27 | SLC25A3 | 19644 | 15268 | -0.522 | -0.7029 | No | ||
| 28 | NDUFS3 | 7553 12683 | 15411 | -0.600 | -0.7055 | No | ||
| 29 | NDUFS8 | 23950 | 15598 | -0.708 | -0.7094 | No | ||
| 30 | ATP5C1 | 8635 | 15756 | -0.827 | -0.7108 | Yes | ||
| 31 | NDUFA2 | 9450 | 15760 | -0.829 | -0.7039 | Yes | ||
| 32 | TIMM9 | 19686 | 15817 | -0.873 | -0.6995 | Yes | ||
| 33 | ATP5J | 880 22555 | 16024 | -1.008 | -0.7020 | Yes | ||
| 34 | UQCRC1 | 10259 | 16148 | -1.088 | -0.6993 | Yes | ||
| 35 | PMPCA | 12421 7350 | 16413 | -1.343 | -0.7021 | Yes | ||
| 36 | ATP5G2 | 12610 | 16579 | -1.540 | -0.6978 | Yes | ||
| 37 | ATP5D | 19949 | 16608 | -1.558 | -0.6860 | Yes | ||
| 38 | NDUFAB1 | 7667 | 16758 | -1.695 | -0.6796 | Yes | ||
| 39 | ATP5G3 | 14558 | 16819 | -1.777 | -0.6676 | Yes | ||
| 40 | ABCB7 | 24085 | 16834 | -1.792 | -0.6531 | Yes | ||
| 41 | SURF1 | 14645 | 16891 | -1.873 | -0.6401 | Yes | ||
| 42 | ATP5O | 22539 | 17106 | -2.180 | -0.6330 | Yes | ||
| 43 | MPV17 | 9407 3505 3543 | 17124 | -2.202 | -0.6152 | Yes | ||
| 44 | ATP5E | 14321 | 17201 | -2.299 | -0.5996 | Yes | ||
| 45 | ATP5A1 | 23505 | 17266 | -2.447 | -0.5822 | Yes | ||
| 46 | NDUFS7 | 19946 | 17484 | -2.874 | -0.5693 | Yes | ||
| 47 | NDUFS2 | 4089 13760 | 17608 | -3.085 | -0.5496 | Yes | ||
| 48 | COX15 | 5931 | 17718 | -3.365 | -0.5268 | Yes | ||
| 49 | NDUFA9 | 12288 | 17848 | -3.704 | -0.5021 | Yes | ||
| 50 | TIMM10 | 11390 2158 | 17944 | -4.026 | -0.4728 | Yes | ||
| 51 | SDHD | 19125 | 18022 | -4.265 | -0.4406 | Yes | ||
| 52 | NDUFV1 | 23953 | 18063 | -4.450 | -0.4047 | Yes | ||
| 53 | TIMM8B | 11391 | 18205 | -4.967 | -0.3699 | Yes | ||
| 54 | BCS1L | 14223 | 18210 | -4.972 | -0.3277 | Yes | ||
| 55 | TIMM50 | 17911 | 18220 | -5.003 | -0.2855 | Yes | ||
| 56 | ATP5G1 | 8636 | 18339 | -5.580 | -0.2442 | Yes | ||
| 57 | TIMM17A | 13822 | 18343 | -5.588 | -0.1967 | Yes | ||
| 58 | PHB2 | 17286 | 18507 | -7.035 | -0.1454 | Yes | ||
| 59 | SLC25A15 | 356 3819 | 18563 | -8.073 | -0.0794 | Yes | ||
| 60 | TIMM13 | 14974 | 18605 | -9.631 | 0.0006 | Yes |