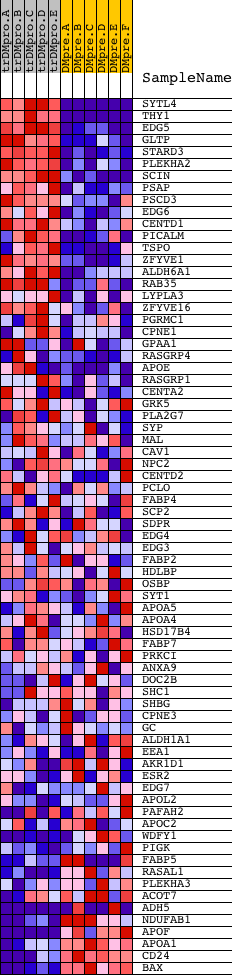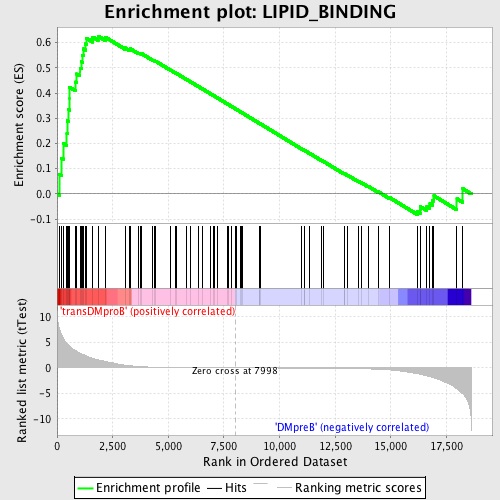
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | LIPID_BINDING |
| Enrichment Score (ES) | 0.6255801 |
| Normalized Enrichment Score (NES) | 1.5259657 |
| Nominal p-value | 0.0021097045 |
| FDR q-value | 0.41445175 |
| FWER p-Value | 0.997 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SYTL4 | 24067 | 93 | 7.857 | 0.0788 | Yes | ||
| 2 | THY1 | 19481 | 201 | 6.463 | 0.1420 | Yes | ||
| 3 | EDG5 | 19216 | 275 | 5.817 | 0.2001 | Yes | ||
| 4 | GLTP | 16419 | 442 | 4.720 | 0.2415 | Yes | ||
| 5 | STARD3 | 20675 | 462 | 4.623 | 0.2898 | Yes | ||
| 6 | PLEKHA2 | 18908 | 524 | 4.401 | 0.3335 | Yes | ||
| 7 | SCIN | 21079 | 556 | 4.289 | 0.3776 | Yes | ||
| 8 | PSAP | 20014 | 559 | 4.274 | 0.4230 | Yes | ||
| 9 | PSCD3 | 16633 | 837 | 3.341 | 0.4438 | Yes | ||
| 10 | EDG6 | 19672 | 886 | 3.222 | 0.4755 | Yes | ||
| 11 | CENTD1 | 16529 | 1028 | 2.872 | 0.4986 | Yes | ||
| 12 | PICALM | 18191 | 1089 | 2.731 | 0.5245 | Yes | ||
| 13 | TSPO | 2204 22408 | 1149 | 2.616 | 0.5492 | Yes | ||
| 14 | ZFYVE1 | 10138 | 1182 | 2.559 | 0.5748 | Yes | ||
| 15 | ALDH6A1 | 21025 | 1280 | 2.351 | 0.5946 | Yes | ||
| 16 | RAB35 | 13386 | 1317 | 2.290 | 0.6171 | Yes | ||
| 17 | LYPLA3 | 18482 | 1599 | 1.865 | 0.6219 | Yes | ||
| 18 | ZFYVE16 | 21395 | 1843 | 1.576 | 0.6256 | Yes | ||
| 19 | PGRMC1 | 24359 | 2180 | 1.240 | 0.6207 | No | ||
| 20 | CPNE1 | 11186 | 3061 | 0.519 | 0.5788 | No | ||
| 21 | GPAA1 | 22446 | 3274 | 0.416 | 0.5718 | No | ||
| 22 | RASGRP4 | 6119 | 3284 | 0.411 | 0.5757 | No | ||
| 23 | APOE | 8618 | 3661 | 0.267 | 0.5582 | No | ||
| 24 | RASGRP1 | 5360 14476 9694 | 3731 | 0.247 | 0.5572 | No | ||
| 25 | CENTA2 | 20748 | 3773 | 0.237 | 0.5575 | No | ||
| 26 | GRK5 | 9036 23814 | 4307 | 0.133 | 0.5302 | No | ||
| 27 | PLA2G7 | 23233 | 4367 | 0.123 | 0.5283 | No | ||
| 28 | SYP | 9958 | 4425 | 0.116 | 0.5265 | No | ||
| 29 | MAL | 2760 14437 | 5101 | 0.063 | 0.4907 | No | ||
| 30 | CAV1 | 8698 | 5321 | 0.053 | 0.4795 | No | ||
| 31 | NPC2 | 21023 | 5370 | 0.051 | 0.4774 | No | ||
| 32 | CENTD2 | 1758 18169 2284 | 5821 | 0.037 | 0.4536 | No | ||
| 33 | PCLO | 11207 6495 | 5975 | 0.033 | 0.4457 | No | ||
| 34 | FABP4 | 8601 | 6365 | 0.025 | 0.4250 | No | ||
| 35 | SCP2 | 5415 2422 9785 5416 | 6517 | 0.022 | 0.4171 | No | ||
| 36 | SDPR | 14252 | 6911 | 0.015 | 0.3960 | No | ||
| 37 | EDG4 | 18595 12005 | 7016 | 0.014 | 0.3906 | No | ||
| 38 | EDG3 | 21639 | 7059 | 0.013 | 0.3884 | No | ||
| 39 | FABP2 | 15430 | 7197 | 0.011 | 0.3812 | No | ||
| 40 | HDLBP | 4017 13873 | 7644 | 0.005 | 0.3572 | No | ||
| 41 | OSBP | 4278 | 7691 | 0.004 | 0.3547 | No | ||
| 42 | SYT1 | 5565 | 7856 | 0.002 | 0.3459 | No | ||
| 43 | APOA5 | 19469 | 8039 | -0.000 | 0.3361 | No | ||
| 44 | APOA4 | 4401 | 8053 | -0.001 | 0.3354 | No | ||
| 45 | HSD17B4 | 23560 | 8252 | -0.003 | 0.3248 | No | ||
| 46 | FABP7 | 20020 | 8273 | -0.004 | 0.3237 | No | ||
| 47 | PRKCI | 9576 | 8313 | -0.004 | 0.3217 | No | ||
| 48 | ANXA9 | 12964 | 9112 | -0.015 | 0.2788 | No | ||
| 49 | DOC2B | 20347 8859 | 9117 | -0.015 | 0.2787 | No | ||
| 50 | SHC1 | 9813 9812 5430 | 9137 | -0.016 | 0.2779 | No | ||
| 51 | SHBG | 20388 | 10974 | -0.046 | 0.1794 | No | ||
| 52 | CPNE3 | 7689 | 11112 | -0.049 | 0.1725 | No | ||
| 53 | GC | 16486 | 11121 | -0.049 | 0.1726 | No | ||
| 54 | ALDH1A1 | 8569 | 11359 | -0.054 | 0.1604 | No | ||
| 55 | EEA1 | 5695 10097 | 11864 | -0.068 | 0.1339 | No | ||
| 56 | AKR1D1 | 17483 | 11954 | -0.071 | 0.1299 | No | ||
| 57 | ESR2 | 21038 | 12895 | -0.112 | 0.0804 | No | ||
| 58 | EDG7 | 15395 | 12922 | -0.115 | 0.0802 | No | ||
| 59 | APOL2 | 22229 | 13056 | -0.122 | 0.0743 | No | ||
| 60 | PAFAH2 | 16045 2429 2483 | 13534 | -0.159 | 0.0503 | No | ||
| 61 | APOC2 | 8616 | 13692 | -0.174 | 0.0437 | No | ||
| 62 | WDFY1 | 7617 | 13989 | -0.205 | 0.0299 | No | ||
| 63 | PIGK | 1815 6834 11629 | 14435 | -0.271 | 0.0088 | No | ||
| 64 | FABP5 | 15637 4958 | 14919 | -0.389 | -0.0131 | No | ||
| 65 | RASAL1 | 16719 | 16187 | -1.126 | -0.0694 | No | ||
| 66 | PLEKHA3 | 14978 | 16319 | -1.245 | -0.0632 | No | ||
| 67 | ACOT7 | 2546 15982 2320 | 16324 | -1.248 | -0.0501 | No | ||
| 68 | ADH5 | 8554 15404 | 16621 | -1.570 | -0.0493 | No | ||
| 69 | NDUFAB1 | 7667 | 16758 | -1.695 | -0.0386 | No | ||
| 70 | APOF | 19843 | 16885 | -1.869 | -0.0254 | No | ||
| 71 | APOA1 | 8614 | 16938 | -1.944 | -0.0075 | No | ||
| 72 | CD24 | 8711 | 17973 | -4.116 | -0.0193 | No | ||
| 73 | BAX | 17832 | 18237 | -5.057 | 0.0204 | No |