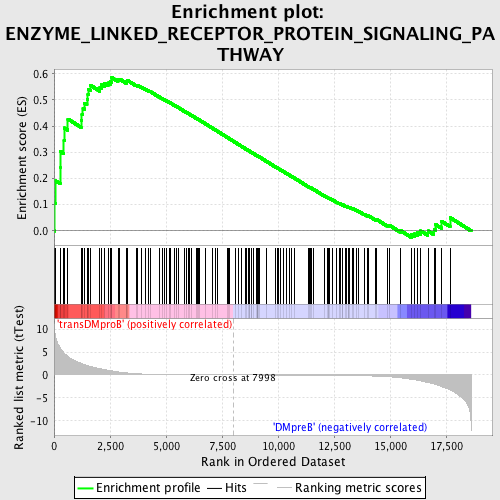
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | ENZYME_LINKED_RECEPTOR_PROTEIN_SIGNALING_PATHWAY |
| Enrichment Score (ES) | 0.5855382 |
| Normalized Enrichment Score (NES) | 1.5315446 |
| Nominal p-value | 0.006355932 |
| FDR q-value | 0.4078667 |
| FWER p-Value | 0.997 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GRB7 | 20673 | 14 | 10.160 | 0.1067 | Yes | ||
| 2 | ENG | 4667 | 66 | 8.260 | 0.1913 | Yes | ||
| 3 | SMAD5 | 21621 | 269 | 5.871 | 0.2425 | Yes | ||
| 4 | GAB1 | 18828 | 276 | 5.806 | 0.3036 | Yes | ||
| 5 | SMAD3 | 19084 | 440 | 4.748 | 0.3450 | Yes | ||
| 6 | PDPK1 | 23097 | 458 | 4.644 | 0.3932 | Yes | ||
| 7 | TGFB1 | 18332 | 616 | 4.024 | 0.4273 | Yes | ||
| 8 | NCK2 | 9448 | 1208 | 2.518 | 0.4219 | Yes | ||
| 9 | SMAD4 | 5058 | 1242 | 2.437 | 0.4459 | Yes | ||
| 10 | ABI1 | 4300 | 1288 | 2.340 | 0.4682 | Yes | ||
| 11 | LEFTY1 | 8877 | 1356 | 2.217 | 0.4881 | Yes | ||
| 12 | FIBP | 7195 | 1469 | 2.036 | 0.5035 | Yes | ||
| 13 | PTK2B | 21776 | 1509 | 1.984 | 0.5224 | Yes | ||
| 14 | GRB2 | 20149 | 1534 | 1.943 | 0.5417 | Yes | ||
| 15 | ACVR1 | 4334 | 1627 | 1.818 | 0.5559 | Yes | ||
| 16 | EGF | 15169 | 2044 | 1.359 | 0.5478 | Yes | ||
| 17 | CDKN1C | 17546 | 2111 | 1.301 | 0.5580 | Yes | ||
| 18 | FNTA | 18904 | 2254 | 1.160 | 0.5626 | Yes | ||
| 19 | PICK1 | 9560 22424 | 2409 | 1.020 | 0.5651 | Yes | ||
| 20 | AP3S1 | 8604 | 2504 | 0.965 | 0.5702 | Yes | ||
| 21 | IGF1R | 9157 | 2572 | 0.900 | 0.5761 | Yes | ||
| 22 | HIPK2 | 17180 | 2574 | 0.899 | 0.5855 | Yes | ||
| 23 | ACVR1B | 4335 22353 | 2853 | 0.647 | 0.5774 | No | ||
| 24 | MAP3K7 | 16255 | 2925 | 0.600 | 0.5799 | No | ||
| 25 | GFRA2 | 21965 | 3223 | 0.439 | 0.5684 | No | ||
| 26 | ZFYVE9 | 10501 2449 | 3239 | 0.432 | 0.5722 | No | ||
| 27 | SNF1LK2 | 6168 10649 | 3260 | 0.419 | 0.5755 | No | ||
| 28 | ACVR2B | 19276 | 3662 | 0.266 | 0.5567 | No | ||
| 29 | SMAD2 | 23511 | 3741 | 0.245 | 0.5551 | No | ||
| 30 | GDF9 | 20887 | 3908 | 0.202 | 0.5482 | No | ||
| 31 | ACVR2A | 8542 | 4076 | 0.167 | 0.5410 | No | ||
| 32 | BMPR2 | 14241 4081 | 4206 | 0.147 | 0.5355 | No | ||
| 33 | CIDEA | 23529 | 4299 | 0.134 | 0.5320 | No | ||
| 34 | ERBB2IP | 3232 12234 | 4690 | 0.088 | 0.5118 | No | ||
| 35 | PXN | 5339 3573 | 4837 | 0.077 | 0.5047 | No | ||
| 36 | PIK3R1 | 3170 | 4927 | 0.072 | 0.5007 | No | ||
| 37 | SHC3 | 21465 | 5010 | 0.068 | 0.4969 | No | ||
| 38 | ACVRL1 | 22354 | 5145 | 0.061 | 0.4903 | No | ||
| 39 | RAPGEF1 | 4218 2860 | 5176 | 0.059 | 0.4893 | No | ||
| 40 | TGFBR1 | 16213 10165 5745 | 5354 | 0.052 | 0.4803 | No | ||
| 41 | PTPRF | 5331 2545 | 5442 | 0.048 | 0.4761 | No | ||
| 42 | TGFBR3 | 16453 | 5532 | 0.045 | 0.4718 | No | ||
| 43 | KLF10 | 22308 | 5836 | 0.037 | 0.4558 | No | ||
| 44 | BDKRB2 | 21163 | 5909 | 0.035 | 0.4523 | No | ||
| 45 | DOK1 | 17104 1018 1177 | 5992 | 0.033 | 0.4482 | No | ||
| 46 | EPGN | 16798 | 6036 | 0.032 | 0.4462 | No | ||
| 47 | SOST | 20210 | 6128 | 0.030 | 0.4416 | No | ||
| 48 | ERBB3 | 19593 | 6360 | 0.025 | 0.4294 | No | ||
| 49 | PTN | 5323 | 6396 | 0.025 | 0.4277 | No | ||
| 50 | BAIAP2 | 449 4236 | 6429 | 0.024 | 0.4262 | No | ||
| 51 | GP6 | 17990 | 6485 | 0.023 | 0.4235 | No | ||
| 52 | GUCA1B | 23211 | 6744 | 0.018 | 0.4097 | No | ||
| 53 | EREG | 4679 16797 | 7084 | 0.013 | 0.3915 | No | ||
| 54 | MUSK | 5199 | 7219 | 0.011 | 0.3844 | No | ||
| 55 | PTPRT | 5333 | 7294 | 0.009 | 0.3805 | No | ||
| 56 | PTPRU | 15738 | 7724 | 0.003 | 0.3573 | No | ||
| 57 | MTSS1 | 5585 22285 | 7799 | 0.003 | 0.3534 | No | ||
| 58 | FOXC2 | 18442 | 7841 | 0.002 | 0.3512 | No | ||
| 59 | FGF5 | 16785 | 8104 | -0.001 | 0.3370 | No | ||
| 60 | GUCY2F | 24045 | 8251 | -0.003 | 0.3292 | No | ||
| 61 | CNKSR1 | 15719 20331 | 8386 | -0.005 | 0.3220 | No | ||
| 62 | RPS6KA5 | 2076 21005 | 8560 | -0.007 | 0.3127 | No | ||
| 63 | ROR1 | 6472 | 8593 | -0.008 | 0.3110 | No | ||
| 64 | OTX2 | 9519 | 8660 | -0.009 | 0.3076 | No | ||
| 65 | MPZL1 | 13777 | 8702 | -0.010 | 0.3055 | No | ||
| 66 | FMOD | 14133 | 8740 | -0.010 | 0.3036 | No | ||
| 67 | NTRK2 | 9491 | 8794 | -0.011 | 0.3008 | No | ||
| 68 | CD3E | 8714 | 8819 | -0.011 | 0.2996 | No | ||
| 69 | SMAD7 | 23513 | 8883 | -0.012 | 0.2963 | No | ||
| 70 | EDN2 | 16102 | 9046 | -0.014 | 0.2877 | No | ||
| 71 | RGMB | 12736 7585 | 9058 | -0.015 | 0.2873 | No | ||
| 72 | CBLC | 17938 | 9067 | -0.015 | 0.2870 | No | ||
| 73 | GUCY2C | 1140 16954 | 9096 | -0.015 | 0.2857 | No | ||
| 74 | SHC1 | 9813 9812 5430 | 9137 | -0.016 | 0.2837 | No | ||
| 75 | IL17F | 13992 | 9150 | -0.016 | 0.2832 | No | ||
| 76 | FRS3 | 23209 | 9497 | -0.021 | 0.2647 | No | ||
| 77 | TDGF1 | 5704 10108 | 9873 | -0.026 | 0.2447 | No | ||
| 78 | GDF5 | 4768 | 9949 | -0.028 | 0.2409 | No | ||
| 79 | IRS1 | 4925 | 10031 | -0.029 | 0.2369 | No | ||
| 80 | KDR | 16509 | 10088 | -0.030 | 0.2341 | No | ||
| 81 | TGFA | 17381 | 10239 | -0.033 | 0.2264 | No | ||
| 82 | LEFTY2 | 6735 | 10359 | -0.035 | 0.2203 | No | ||
| 83 | FOXH1 | 22238 | 10491 | -0.037 | 0.2136 | No | ||
| 84 | CITED2 | 5118 14477 | 10616 | -0.039 | 0.2073 | No | ||
| 85 | REPS2 | 24018 | 10743 | -0.041 | 0.2009 | No | ||
| 86 | SRC | 5507 | 11343 | -0.054 | 0.1691 | No | ||
| 87 | SMURF1 | 13289 7989 | 11408 | -0.055 | 0.1662 | No | ||
| 88 | BMPR1A | 8660 4450 | 11434 | -0.056 | 0.1655 | No | ||
| 89 | PTPRD | 11554 5330 | 11472 | -0.056 | 0.1641 | No | ||
| 90 | GRB10 | 4799 | 11590 | -0.060 | 0.1584 | No | ||
| 91 | FGFR3 | 8969 3566 | 12064 | -0.074 | 0.1336 | No | ||
| 92 | TEK | 16175 | 12187 | -0.079 | 0.1278 | No | ||
| 93 | IGF2 | 17550 | 12252 | -0.082 | 0.1252 | No | ||
| 94 | EFNB3 | 20391 | 12263 | -0.082 | 0.1255 | No | ||
| 95 | PTPRG | 22097 3151 | 12308 | -0.085 | 0.1241 | No | ||
| 96 | FLT3 | 16288 | 12437 | -0.091 | 0.1181 | No | ||
| 97 | EGFR | 1329 20944 | 12619 | -0.098 | 0.1093 | No | ||
| 98 | FGFR4 | 3226 | 12719 | -0.103 | 0.1051 | No | ||
| 99 | GDF10 | 4767 | 12772 | -0.106 | 0.1034 | No | ||
| 100 | MAPK8IP2 | 77 | 12884 | -0.112 | 0.0986 | No | ||
| 101 | NTRK3 | 2069 3652 3705 9492 | 12888 | -0.112 | 0.0996 | No | ||
| 102 | GDF15 | 10721 | 13018 | -0.120 | 0.0939 | No | ||
| 103 | ERBB2 | 8913 | 13054 | -0.122 | 0.0933 | No | ||
| 104 | SNX6 | 21070 | 13131 | -0.127 | 0.0905 | No | ||
| 105 | FLT4 | 20904 1461 | 13146 | -0.128 | 0.0911 | No | ||
| 106 | AKT1 | 8568 | 13190 | -0.131 | 0.0902 | No | ||
| 107 | EID2 | 11815 | 13306 | -0.140 | 0.0854 | No | ||
| 108 | NRTN | 22923 | 13352 | -0.142 | 0.0845 | No | ||
| 109 | HPGD | 4867 | 13497 | -0.156 | 0.0784 | No | ||
| 110 | PTPRJ | 9664 | 13584 | -0.163 | 0.0755 | No | ||
| 111 | CBL | 19154 | 13840 | -0.189 | 0.0637 | No | ||
| 112 | INSR | 18950 | 14009 | -0.207 | 0.0568 | No | ||
| 113 | NTRK1 | 15299 | 14033 | -0.210 | 0.0577 | No | ||
| 114 | LCP2 | 4988 9268 | 14367 | -0.258 | 0.0425 | No | ||
| 115 | CDH13 | 4506 3826 | 14398 | -0.263 | 0.0436 | No | ||
| 116 | SMAD1 | 18831 922 | 14865 | -0.374 | 0.0224 | No | ||
| 117 | PRLR | 9620 5291 | 14991 | -0.411 | 0.0200 | No | ||
| 118 | STUB1 | 23070 | 15472 | -0.640 | 0.0008 | No | ||
| 119 | LTBP2 | 5007 | 15951 | -0.981 | -0.0147 | No | ||
| 120 | EPS15 | 4677 | 16102 | -1.058 | -0.0116 | No | ||
| 121 | ADRB2 | 23422 | 16221 | -1.155 | -0.0058 | No | ||
| 122 | BCAR1 | 18741 | 16371 | -1.292 | -0.0002 | No | ||
| 123 | EPS8 | 16948 | 16690 | -1.625 | -0.0002 | No | ||
| 124 | CDKN2B | 15840 | 16958 | -1.965 | 0.0062 | No | ||
| 125 | SOCS1 | 4522 | 17035 | -2.077 | 0.0240 | No | ||
| 126 | AGRN | 4361 | 17303 | -2.527 | 0.0363 | No | ||
| 127 | PIK3R3 | 5248 | 17678 | -3.275 | 0.0507 | No |

