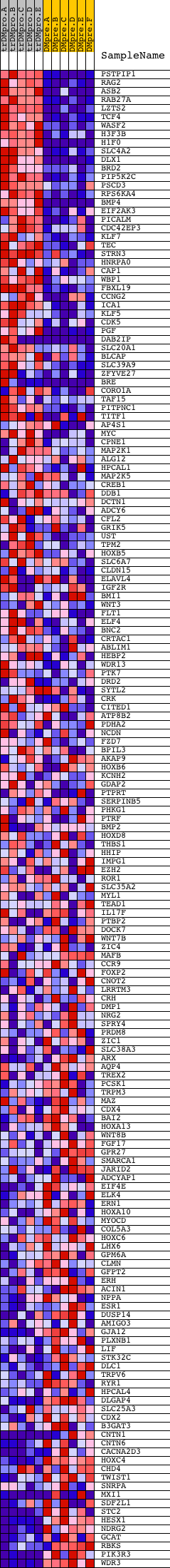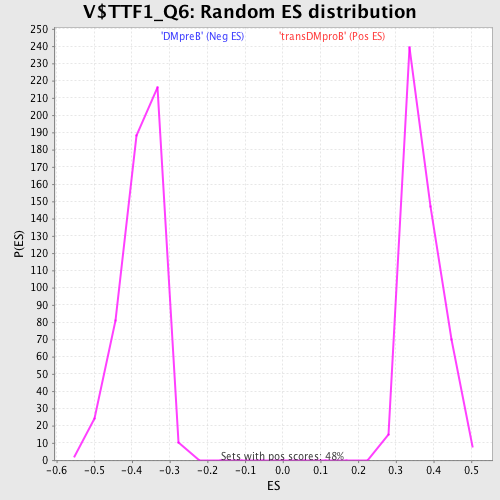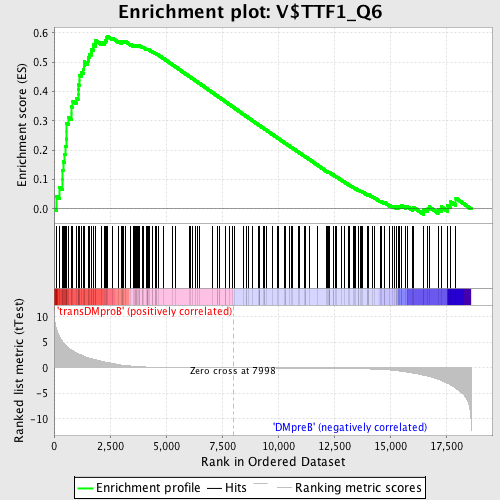
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$TTF1_Q6 |
| Enrichment Score (ES) | 0.5884401 |
| Normalized Enrichment Score (NES) | 1.5937177 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.051193494 |
| FWER p-Value | 0.347 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSTPIP1 | 19437 | 104 | 7.723 | 0.0428 | Yes | ||
| 2 | RAG2 | 14941 | 247 | 6.107 | 0.0733 | Yes | ||
| 3 | ASB2 | 20996 | 363 | 5.184 | 0.0996 | Yes | ||
| 4 | RAB27A | 19381 | 367 | 5.170 | 0.1318 | Yes | ||
| 5 | LZTS2 | 10362 | 401 | 4.974 | 0.1612 | Yes | ||
| 6 | TCF4 | 5642 | 463 | 4.620 | 0.1868 | Yes | ||
| 7 | WASF2 | 6326 | 492 | 4.509 | 0.2136 | Yes | ||
| 8 | H3F3B | 4839 | 539 | 4.344 | 0.2383 | Yes | ||
| 9 | H1F0 | 22428 | 551 | 4.314 | 0.2647 | Yes | ||
| 10 | SLC4A2 | 9828 | 552 | 4.310 | 0.2917 | Yes | ||
| 11 | DLX1 | 14988 | 648 | 3.891 | 0.3110 | Yes | ||
| 12 | BRD2 | 4735 8984 7628 | 771 | 3.522 | 0.3264 | Yes | ||
| 13 | PIP5K2C | 19606 | 772 | 3.521 | 0.3485 | Yes | ||
| 14 | PSCD3 | 16633 | 837 | 3.341 | 0.3659 | Yes | ||
| 15 | RPS6KA4 | 23804 | 985 | 2.977 | 0.3766 | Yes | ||
| 16 | BMP4 | 21863 | 1070 | 2.788 | 0.3895 | Yes | ||
| 17 | EIF2AK3 | 17421 | 1074 | 2.775 | 0.4068 | Yes | ||
| 18 | PICALM | 18191 | 1089 | 2.731 | 0.4231 | Yes | ||
| 19 | CDC42EP3 | 6436 | 1130 | 2.659 | 0.4376 | Yes | ||
| 20 | KLF7 | 8205 | 1131 | 2.659 | 0.4543 | Yes | ||
| 21 | TEC | 16514 | 1209 | 2.517 | 0.4659 | Yes | ||
| 22 | STRN3 | 884 21074 | 1300 | 2.324 | 0.4756 | Yes | ||
| 23 | HNRPA0 | 13382 | 1334 | 2.257 | 0.4879 | Yes | ||
| 24 | CAP1 | 8684 | 1350 | 2.228 | 0.5011 | Yes | ||
| 25 | WBP1 | 17101 | 1512 | 1.982 | 0.5047 | Yes | ||
| 26 | FBXL19 | 6134 | 1536 | 1.943 | 0.5157 | Yes | ||
| 27 | CCNG2 | 16790 | 1565 | 1.905 | 0.5261 | Yes | ||
| 28 | ICA1 | 17217 | 1650 | 1.788 | 0.5327 | Yes | ||
| 29 | KLF5 | 4456 | 1663 | 1.774 | 0.5432 | Yes | ||
| 30 | CDK5 | 16591 | 1747 | 1.669 | 0.5492 | Yes | ||
| 31 | PGF | 21020 | 1750 | 1.666 | 0.5595 | Yes | ||
| 32 | DAB2IP | 12808 2692 | 1846 | 1.573 | 0.5642 | Yes | ||
| 33 | SLC20A1 | 14855 | 1856 | 1.563 | 0.5735 | Yes | ||
| 34 | BLCAP | 14371 | 2125 | 1.289 | 0.5670 | Yes | ||
| 35 | SLC39A9 | 11597 | 2240 | 1.172 | 0.5682 | Yes | ||
| 36 | ZFYVE27 | 23849 | 2283 | 1.133 | 0.5730 | Yes | ||
| 37 | BRE | 3640 4226 16879 | 2323 | 1.096 | 0.5778 | Yes | ||
| 38 | CORO1A | 1066 17631 1074 | 2352 | 1.079 | 0.5830 | Yes | ||
| 39 | TAF15 | 20731 | 2375 | 1.055 | 0.5884 | Yes | ||
| 40 | PITPNC1 | 7759 | 2612 | 0.862 | 0.5810 | No | ||
| 41 | TITF1 | 10180 21062 | 2893 | 0.618 | 0.5697 | No | ||
| 42 | AP4S1 | 21276 | 2992 | 0.557 | 0.5679 | No | ||
| 43 | MYC | 22465 9435 | 3045 | 0.531 | 0.5684 | No | ||
| 44 | CPNE1 | 11186 | 3061 | 0.519 | 0.5709 | No | ||
| 45 | MAP2K1 | 19082 | 3106 | 0.498 | 0.5716 | No | ||
| 46 | ALG12 | 22167 | 3208 | 0.447 | 0.5689 | No | ||
| 47 | HPCAL1 | 21307 2123 | 3431 | 0.348 | 0.5591 | No | ||
| 48 | MAP2K5 | 19088 | 3538 | 0.306 | 0.5552 | No | ||
| 49 | CREB1 | 3990 8782 4558 4093 | 3581 | 0.288 | 0.5548 | No | ||
| 50 | DDB1 | 23931 | 3636 | 0.274 | 0.5535 | No | ||
| 51 | DCTN1 | 1100 17392 | 3645 | 0.272 | 0.5548 | No | ||
| 52 | ADCY6 | 22139 2283 8551 | 3656 | 0.270 | 0.5560 | No | ||
| 53 | CFL2 | 2155 21069 | 3697 | 0.257 | 0.5554 | No | ||
| 54 | GRIK5 | 350 17930 | 3733 | 0.247 | 0.5551 | No | ||
| 55 | UST | 6865 | 3754 | 0.241 | 0.5555 | No | ||
| 56 | TPM2 | 2367 2437 | 3799 | 0.229 | 0.5545 | No | ||
| 57 | HOXB5 | 20687 | 3804 | 0.229 | 0.5558 | No | ||
| 58 | SLC6A7 | 10772 | 3928 | 0.197 | 0.5503 | No | ||
| 59 | CLDN15 | 16667 3572 | 3965 | 0.188 | 0.5495 | No | ||
| 60 | ELAVL4 | 15805 4889 9137 | 3984 | 0.185 | 0.5497 | No | ||
| 61 | IGF2R | 23123 | 4111 | 0.162 | 0.5439 | No | ||
| 62 | BMI1 | 4448 15113 | 4154 | 0.154 | 0.5426 | No | ||
| 63 | WNT3 | 20635 | 4171 | 0.151 | 0.5427 | No | ||
| 64 | FLT1 | 3483 16287 | 4189 | 0.149 | 0.5427 | No | ||
| 65 | ELF4 | 24162 | 4199 | 0.148 | 0.5431 | No | ||
| 66 | BNC2 | 15850 | 4247 | 0.142 | 0.5415 | No | ||
| 67 | CRTAC1 | 23673 | 4402 | 0.119 | 0.5339 | No | ||
| 68 | ABLIM1 | 3717 3766 3679 | 4408 | 0.118 | 0.5343 | No | ||
| 69 | HEBP2 | 19812 | 4506 | 0.106 | 0.5297 | No | ||
| 70 | WDR13 | 7859 13112 | 4510 | 0.106 | 0.5302 | No | ||
| 71 | PTK7 | 22959 | 4569 | 0.098 | 0.5277 | No | ||
| 72 | DRD2 | 19461 | 4591 | 0.096 | 0.5272 | No | ||
| 73 | SYTL2 | 18190 1539 3730 | 4681 | 0.088 | 0.5229 | No | ||
| 74 | CRK | 4559 1249 | 4878 | 0.075 | 0.5127 | No | ||
| 75 | CITED1 | 24091 | 5264 | 0.056 | 0.4922 | No | ||
| 76 | ATP8B2 | 12055 | 5417 | 0.049 | 0.4843 | No | ||
| 77 | PDHA2 | 15153 | 6037 | 0.032 | 0.4509 | No | ||
| 78 | NCDN | 15756 2487 6471 | 6105 | 0.030 | 0.4475 | No | ||
| 79 | FZD7 | 14242 | 6155 | 0.029 | 0.4450 | No | ||
| 80 | BPIL3 | 10459 | 6304 | 0.026 | 0.4371 | No | ||
| 81 | AKAP9 | 16604 16603 4150 8303 | 6422 | 0.024 | 0.4309 | No | ||
| 82 | HOXB6 | 20688 | 6473 | 0.023 | 0.4284 | No | ||
| 83 | KCNH2 | 16592 | 7060 | 0.013 | 0.3967 | No | ||
| 84 | GDAP2 | 15481 | 7291 | 0.009 | 0.3843 | No | ||
| 85 | PTPRT | 5333 | 7294 | 0.009 | 0.3842 | No | ||
| 86 | SERPINB5 | 9861 | 7374 | 0.008 | 0.3800 | No | ||
| 87 | PHKG1 | 16361 | 7397 | 0.008 | 0.3788 | No | ||
| 88 | PTRF | 9667 | 7657 | 0.004 | 0.3648 | No | ||
| 89 | BMP2 | 14833 | 7661 | 0.004 | 0.3647 | No | ||
| 90 | HOXD8 | 9116 | 7822 | 0.002 | 0.3560 | No | ||
| 91 | THBS1 | 5748 14910 | 7978 | 0.000 | 0.3476 | No | ||
| 92 | HHIP | 4849 | 8030 | -0.000 | 0.3449 | No | ||
| 93 | IMPG1 | 19052 | 8448 | -0.006 | 0.3223 | No | ||
| 94 | EZH2 | 1092 17163 | 8583 | -0.008 | 0.3151 | No | ||
| 95 | ROR1 | 6472 | 8593 | -0.008 | 0.3146 | No | ||
| 96 | SLC35A2 | 24389 | 8674 | -0.009 | 0.3103 | No | ||
| 97 | MYL1 | 4015 | 8866 | -0.012 | 0.3001 | No | ||
| 98 | TEAD1 | 18121 | 9106 | -0.015 | 0.2872 | No | ||
| 99 | IL17F | 13992 | 9150 | -0.016 | 0.2850 | No | ||
| 100 | PTBP2 | 7074 | 9163 | -0.016 | 0.2844 | No | ||
| 101 | DOCK7 | 12506 | 9177 | -0.016 | 0.2838 | No | ||
| 102 | WNT7B | 22172 | 9361 | -0.019 | 0.2740 | No | ||
| 103 | ZIC4 | 5992 | 9402 | -0.019 | 0.2720 | No | ||
| 104 | MAFB | 14367 | 9410 | -0.019 | 0.2717 | No | ||
| 105 | CCR9 | 8753 | 9483 | -0.021 | 0.2679 | No | ||
| 106 | FOXP2 | 4312 17523 8523 | 9742 | -0.024 | 0.2541 | No | ||
| 107 | CNOT2 | 12992 3332 453 3357 | 9981 | -0.028 | 0.2414 | No | ||
| 108 | LRRTM3 | 19744 | 10007 | -0.029 | 0.2402 | No | ||
| 109 | CRH | 8784 | 10293 | -0.034 | 0.2249 | No | ||
| 110 | DMP1 | 16776 | 10319 | -0.034 | 0.2238 | No | ||
| 111 | NRG2 | 4274 | 10326 | -0.034 | 0.2237 | No | ||
| 112 | SPRY4 | 23448 | 10528 | -0.037 | 0.2130 | No | ||
| 113 | PRDM8 | 8084 | 10613 | -0.039 | 0.2087 | No | ||
| 114 | ZIC1 | 19040 | 10649 | -0.039 | 0.2071 | No | ||
| 115 | SLC38A3 | 13319 | 10929 | -0.045 | 0.1922 | No | ||
| 116 | ARX | 24290 | 10964 | -0.045 | 0.1907 | No | ||
| 117 | AQP4 | 4403 | 11185 | -0.050 | 0.1790 | No | ||
| 118 | TREX2 | 24137 | 11205 | -0.051 | 0.1783 | No | ||
| 119 | PCSK1 | 21600 | 11208 | -0.051 | 0.1785 | No | ||
| 120 | TRPM3 | 23904 10357 5927 | 11416 | -0.055 | 0.1677 | No | ||
| 121 | MAZ | 1327 17623 | 11744 | -0.064 | 0.1503 | No | ||
| 122 | CDX4 | 24273 | 12165 | -0.078 | 0.1280 | No | ||
| 123 | BAI2 | 2541 2379 16067 | 12224 | -0.080 | 0.1254 | No | ||
| 124 | HOXA13 | 9107 | 12257 | -0.082 | 0.1242 | No | ||
| 125 | WNT8B | 23841 | 12271 | -0.083 | 0.1240 | No | ||
| 126 | FGF17 | 21757 | 12274 | -0.083 | 0.1244 | No | ||
| 127 | GPR27 | 13703 | 12292 | -0.084 | 0.1240 | No | ||
| 128 | SMARCA1 | 24165 | 12455 | -0.091 | 0.1158 | No | ||
| 129 | JARID2 | 9200 | 12466 | -0.092 | 0.1158 | No | ||
| 130 | ADCYAP1 | 4348 | 12557 | -0.095 | 0.1115 | No | ||
| 131 | EIF4E | 15403 1827 8890 | 12621 | -0.098 | 0.1087 | No | ||
| 132 | ELK4 | 8895 4664 3937 | 12844 | -0.110 | 0.0974 | No | ||
| 133 | ERN1 | 8137 | 12954 | -0.116 | 0.0922 | No | ||
| 134 | HOXA10 | 17145 989 | 13155 | -0.129 | 0.0822 | No | ||
| 135 | MYOCD | 1382 20406 1401 | 13194 | -0.132 | 0.0809 | No | ||
| 136 | COL5A3 | 19219 | 13348 | -0.142 | 0.0735 | No | ||
| 137 | HOXC6 | 22339 2302 | 13427 | -0.148 | 0.0702 | No | ||
| 138 | LHX6 | 9275 2914 | 13474 | -0.153 | 0.0687 | No | ||
| 139 | GPM6A | 3834 10618 | 13606 | -0.166 | 0.0626 | No | ||
| 140 | CLMN | 20988 2119 2104 | 13607 | -0.166 | 0.0637 | No | ||
| 141 | GFPT2 | 4775 | 13684 | -0.173 | 0.0606 | No | ||
| 142 | ERH | 9614 4681 | 13713 | -0.176 | 0.0602 | No | ||
| 143 | ACIN1 | 3319 2807 21832 | 13776 | -0.182 | 0.0580 | No | ||
| 144 | NPPA | 298 | 13969 | -0.202 | 0.0489 | No | ||
| 145 | ESR1 | 20097 4685 | 14010 | -0.207 | 0.0480 | No | ||
| 146 | DUSP14 | 12124 | 14021 | -0.209 | 0.0488 | No | ||
| 147 | AMIGO3 | 19314 | 14039 | -0.211 | 0.0492 | No | ||
| 148 | GJA12 | 20433 1323 | 14213 | -0.236 | 0.0413 | No | ||
| 149 | PLXNB1 | 19301 | 14305 | -0.248 | 0.0379 | No | ||
| 150 | LIF | 956 667 20960 | 14556 | -0.293 | 0.0262 | No | ||
| 151 | STK32C | 17580 | 14618 | -0.311 | 0.0248 | No | ||
| 152 | DLC1 | 11931 3855 | 14744 | -0.344 | 0.0202 | No | ||
| 153 | TRPV6 | 17171 | 14770 | -0.350 | 0.0210 | No | ||
| 154 | RYR1 | 17903 17902 9770 | 14986 | -0.410 | 0.0119 | No | ||
| 155 | HPCAL4 | 16096 | 15119 | -0.462 | 0.0077 | No | ||
| 156 | DLGAP4 | 2950 10461 | 15175 | -0.484 | 0.0077 | No | ||
| 157 | SLC25A3 | 19644 | 15268 | -0.522 | 0.0060 | No | ||
| 158 | CDX2 | 16289 | 15282 | -0.527 | 0.0086 | No | ||
| 159 | B3GAT3 | 23938 | 15383 | -0.578 | 0.0068 | No | ||
| 160 | CNTN1 | 4538 | 15412 | -0.600 | 0.0090 | No | ||
| 161 | CNTN6 | 17346 | 15503 | -0.655 | 0.0083 | No | ||
| 162 | CACNA2D3 | 8677 | 15505 | -0.655 | 0.0123 | No | ||
| 163 | HOXC4 | 4864 | 15695 | -0.780 | 0.0069 | No | ||
| 164 | CHD4 | 8418 17281 4225 | 15754 | -0.826 | 0.0090 | No | ||
| 165 | TWIST1 | 21291 | 16003 | -1.001 | 0.0018 | No | ||
| 166 | SNRPA | 7007 11992 7008 | 16045 | -1.019 | 0.0060 | No | ||
| 167 | MXI1 | 23813 5139 | 16502 | -1.464 | -0.0096 | No | ||
| 168 | SDF2L1 | 22649 | 16509 | -1.471 | -0.0007 | No | ||
| 169 | STC2 | 5526 | 16684 | -1.618 | 0.0000 | No | ||
| 170 | HESX1 | 22070 | 16740 | -1.676 | 0.0075 | No | ||
| 171 | NDRG2 | 21843 | 17140 | -2.219 | -0.0002 | No | ||
| 172 | GCAT | 11234 | 17278 | -2.477 | 0.0079 | No | ||
| 173 | RBKS | 16568 | 17566 | -3.021 | 0.0113 | No | ||
| 174 | PIK3R3 | 5248 | 17678 | -3.275 | 0.0258 | No | ||
| 175 | WDR3 | 15225 | 17936 | -3.998 | 0.0369 | No |