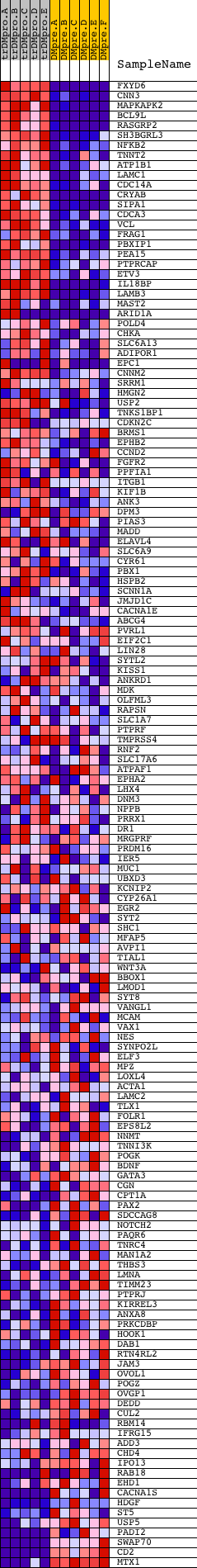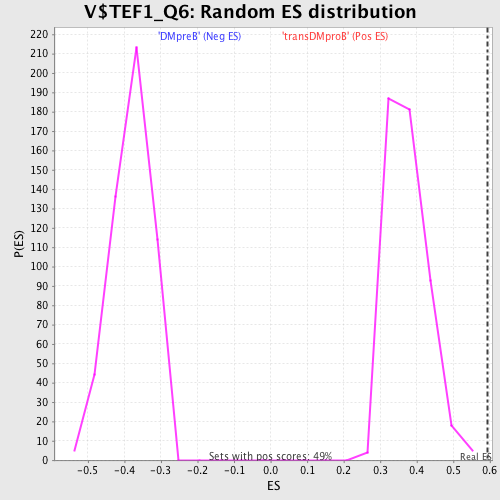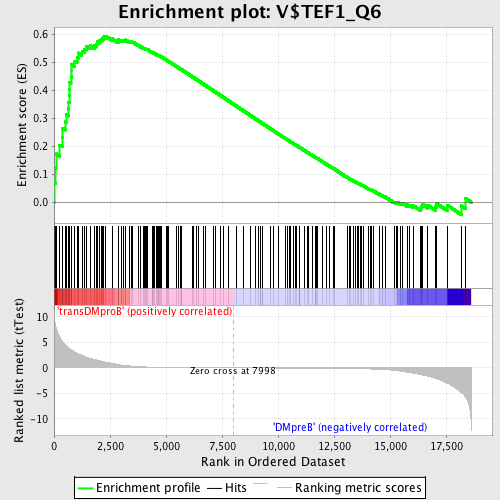
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$TEF1_Q6 |
| Enrichment Score (ES) | 0.5945995 |
| Normalized Enrichment Score (NES) | 1.5843765 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.04868561 |
| FWER p-Value | 0.386 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FXYD6 | 19471 | 4 | 10.857 | 0.0702 | Yes | ||
| 2 | CNN3 | 12983 | 56 | 8.459 | 0.1223 | Yes | ||
| 3 | MAPKAPK2 | 13838 | 85 | 7.980 | 0.1726 | Yes | ||
| 4 | BCL9L | 19475 | 237 | 6.203 | 0.2046 | Yes | ||
| 5 | RASGRP2 | 9693 | 358 | 5.214 | 0.2319 | Yes | ||
| 6 | SH3BGRL3 | 15722 | 369 | 5.157 | 0.2648 | Yes | ||
| 7 | NFKB2 | 23810 | 491 | 4.513 | 0.2876 | Yes | ||
| 8 | TNNT2 | 14113 | 535 | 4.364 | 0.3135 | Yes | ||
| 9 | ATP1B1 | 4420 | 631 | 3.969 | 0.3341 | Yes | ||
| 10 | LAMC1 | 13805 | 653 | 3.867 | 0.3581 | Yes | ||
| 11 | CDC14A | 15184 | 682 | 3.756 | 0.3809 | Yes | ||
| 12 | CRYAB | 19457 | 694 | 3.726 | 0.4045 | Yes | ||
| 13 | SIPA1 | 9820 | 701 | 3.703 | 0.4282 | Yes | ||
| 14 | CDCA3 | 17284 | 754 | 3.560 | 0.4485 | Yes | ||
| 15 | VCL | 22083 | 775 | 3.518 | 0.4702 | Yes | ||
| 16 | FRAG1 | 6129 18165 1724 | 778 | 3.515 | 0.4929 | Yes | ||
| 17 | PBXIP1 | 6028 | 929 | 3.103 | 0.5049 | Yes | ||
| 18 | PEA15 | 13753 | 1023 | 2.877 | 0.5185 | Yes | ||
| 19 | PTPRCAP | 23767 | 1069 | 2.788 | 0.5342 | Yes | ||
| 20 | ETV3 | 11294 | 1244 | 2.434 | 0.5405 | Yes | ||
| 21 | IL18BP | 17730 | 1363 | 2.208 | 0.5485 | Yes | ||
| 22 | LAMB3 | 14007 | 1431 | 2.097 | 0.5584 | Yes | ||
| 23 | MAST2 | 9423 | 1616 | 1.838 | 0.5604 | Yes | ||
| 24 | ARID1A | 8214 2501 13554 | 1797 | 1.633 | 0.5612 | Yes | ||
| 25 | POLD4 | 12822 | 1902 | 1.515 | 0.5654 | Yes | ||
| 26 | CHKA | 23759 | 1924 | 1.488 | 0.5739 | Yes | ||
| 27 | SLC6A13 | 17300 | 2005 | 1.398 | 0.5787 | Yes | ||
| 28 | ADIPOR1 | 14126 | 2095 | 1.315 | 0.5824 | Yes | ||
| 29 | EPC1 | 2039 8907 2033 | 2166 | 1.256 | 0.5867 | Yes | ||
| 30 | CNNM2 | 8251 | 2222 | 1.194 | 0.5915 | Yes | ||
| 31 | SRRM1 | 6958 | 2300 | 1.120 | 0.5946 | Yes | ||
| 32 | HMGN2 | 9095 | 2587 | 0.887 | 0.5849 | No | ||
| 33 | USP2 | 19480 3052 3043 | 2862 | 0.641 | 0.5742 | No | ||
| 34 | TNKS1BP1 | 14970 | 2877 | 0.631 | 0.5775 | No | ||
| 35 | CDKN2C | 15806 2321 | 2881 | 0.629 | 0.5814 | No | ||
| 36 | BRMS1 | 8403 | 3013 | 0.549 | 0.5779 | No | ||
| 37 | EPHB2 | 4675 2440 8910 | 3029 | 0.541 | 0.5806 | No | ||
| 38 | CCND2 | 16987 | 3097 | 0.503 | 0.5802 | No | ||
| 39 | FGFR2 | 1917 4722 1119 | 3175 | 0.460 | 0.5791 | No | ||
| 40 | PPFIA1 | 17538 6136 | 3197 | 0.452 | 0.5808 | No | ||
| 41 | ITGB1 | 3872 18411 | 3355 | 0.377 | 0.5748 | No | ||
| 42 | KIF1B | 15671 2476 4950 15673 | 3373 | 0.371 | 0.5763 | No | ||
| 43 | ANK3 | 8591 3445 3304 3381 | 3439 | 0.346 | 0.5750 | No | ||
| 44 | DPM3 | 15540 | 3505 | 0.319 | 0.5735 | No | ||
| 45 | PIAS3 | 15491 672 1906 | 3768 | 0.238 | 0.5609 | No | ||
| 46 | MADD | 2926 14526 | 3877 | 0.213 | 0.5564 | No | ||
| 47 | ELAVL4 | 15805 4889 9137 | 3984 | 0.185 | 0.5519 | No | ||
| 48 | SLC6A9 | 4783 | 4048 | 0.173 | 0.5496 | No | ||
| 49 | CYR61 | 9159 15145 9311 5024 | 4100 | 0.164 | 0.5479 | No | ||
| 50 | PBX1 | 5224 | 4121 | 0.161 | 0.5479 | No | ||
| 51 | HSPB2 | 19121 | 4150 | 0.155 | 0.5473 | No | ||
| 52 | SCNN1A | 17278 | 4370 | 0.123 | 0.5363 | No | ||
| 53 | JMJD1C | 11531 19996 | 4393 | 0.120 | 0.5359 | No | ||
| 54 | CACNA1E | 4465 | 4423 | 0.116 | 0.5351 | No | ||
| 55 | ABCG4 | 19151 | 4498 | 0.107 | 0.5317 | No | ||
| 56 | PVRL1 | 3006 19482 12211 7192 | 4590 | 0.097 | 0.5274 | No | ||
| 57 | EIF2C1 | 10672 | 4632 | 0.093 | 0.5258 | No | ||
| 58 | LIN28 | 15723 | 4666 | 0.090 | 0.5246 | No | ||
| 59 | SYTL2 | 18190 1539 3730 | 4681 | 0.088 | 0.5244 | No | ||
| 60 | KISS1 | 6603 | 4698 | 0.087 | 0.5241 | No | ||
| 61 | ANKRD1 | 23689 | 4758 | 0.082 | 0.5215 | No | ||
| 62 | MDK | 14523 | 4786 | 0.080 | 0.5205 | No | ||
| 63 | OLFML3 | 15216 | 5031 | 0.066 | 0.5077 | No | ||
| 64 | RAPSN | 2879 14951 | 5075 | 0.064 | 0.5058 | No | ||
| 65 | SLC1A7 | 16147 | 5106 | 0.063 | 0.5046 | No | ||
| 66 | PTPRF | 5331 2545 | 5442 | 0.048 | 0.4868 | No | ||
| 67 | TMPRSS4 | 2981 19138 3049 | 5555 | 0.044 | 0.4810 | No | ||
| 68 | RNF2 | 9728 | 5644 | 0.042 | 0.4765 | No | ||
| 69 | SLC17A6 | 18226 | 5684 | 0.041 | 0.4747 | No | ||
| 70 | ATPAF1 | 16126 | 5700 | 0.041 | 0.4741 | No | ||
| 71 | EPHA2 | 16006 | 6188 | 0.029 | 0.4479 | No | ||
| 72 | LHX4 | 13800 | 6217 | 0.028 | 0.4466 | No | ||
| 73 | DNM3 | 5948 | 6349 | 0.025 | 0.4397 | No | ||
| 74 | NPPB | 15994 | 6427 | 0.024 | 0.4357 | No | ||
| 75 | PRRX1 | 9595 5271 | 6683 | 0.019 | 0.4220 | No | ||
| 76 | DR1 | 16762 | 6756 | 0.018 | 0.4182 | No | ||
| 77 | MRGPRF | 17995 | 7111 | 0.012 | 0.3991 | No | ||
| 78 | PRDM16 | 12893 | 7208 | 0.011 | 0.3940 | No | ||
| 79 | IER5 | 13802 | 7423 | 0.008 | 0.3824 | No | ||
| 80 | MUC1 | 1849 1799 1787 24354 | 7549 | 0.006 | 0.3757 | No | ||
| 81 | UBXD3 | 15701 | 7805 | 0.003 | 0.3619 | No | ||
| 82 | KCNIP2 | 23655 3725 | 8158 | -0.002 | 0.3429 | No | ||
| 83 | CYP26A1 | 23871 3695 | 8466 | -0.006 | 0.3263 | No | ||
| 84 | EGR2 | 8886 | 8783 | -0.011 | 0.3092 | No | ||
| 85 | SYT2 | 14122 | 8998 | -0.014 | 0.2977 | No | ||
| 86 | SHC1 | 9813 9812 5430 | 9137 | -0.016 | 0.2904 | No | ||
| 87 | MFAP5 | 17296 | 9206 | -0.017 | 0.2868 | No | ||
| 88 | AVPI1 | 23675 | 9214 | -0.017 | 0.2865 | No | ||
| 89 | TIAL1 | 996 5753 | 9296 | -0.018 | 0.2822 | No | ||
| 90 | WNT3A | 20431 | 9309 | -0.018 | 0.2817 | No | ||
| 91 | BBOX1 | 5010 9290 | 9680 | -0.023 | 0.2618 | No | ||
| 92 | LMOD1 | 14119 | 9804 | -0.025 | 0.2553 | No | ||
| 93 | SYT8 | 18006 | 10020 | -0.029 | 0.2439 | No | ||
| 94 | VANGL1 | 6034 | 10349 | -0.035 | 0.2263 | No | ||
| 95 | MCAM | 3109 19479 | 10428 | -0.036 | 0.2224 | No | ||
| 96 | VAX1 | 23636 | 10511 | -0.037 | 0.2182 | No | ||
| 97 | NES | 5162 9454 | 10544 | -0.038 | 0.2167 | No | ||
| 98 | SYNPO2L | 7582 | 10671 | -0.040 | 0.2101 | No | ||
| 99 | ELF3 | 8894 13824 | 10790 | -0.042 | 0.2040 | No | ||
| 100 | MPZ | 9408 | 10839 | -0.043 | 0.2017 | No | ||
| 101 | LOXL4 | 23672 | 10932 | -0.045 | 0.1970 | No | ||
| 102 | ACTA1 | 18715 | 10949 | -0.045 | 0.1964 | No | ||
| 103 | LAMC2 | 4983 9266 13806 3975 | 10971 | -0.046 | 0.1956 | No | ||
| 104 | TLX1 | 23839 | 11170 | -0.050 | 0.1852 | No | ||
| 105 | FOLR1 | 17731 | 11298 | -0.053 | 0.1786 | No | ||
| 106 | EPS8L2 | 18012 | 11361 | -0.054 | 0.1756 | No | ||
| 107 | NNMT | 19131 | 11526 | -0.058 | 0.1671 | No | ||
| 108 | TNNI3K | 11495 | 11651 | -0.061 | 0.1608 | No | ||
| 109 | POGK | 7739 | 11666 | -0.062 | 0.1604 | No | ||
| 110 | BDNF | 14926 2797 | 11691 | -0.062 | 0.1595 | No | ||
| 111 | GATA3 | 9004 | 11758 | -0.064 | 0.1564 | No | ||
| 112 | CGN | 12896 12897 7701 | 11975 | -0.072 | 0.1452 | No | ||
| 113 | CPT1A | 23756 | 12142 | -0.077 | 0.1367 | No | ||
| 114 | PAX2 | 9530 24416 | 12304 | -0.085 | 0.1285 | No | ||
| 115 | SDCCAG8 | 14034 | 12474 | -0.092 | 0.1199 | No | ||
| 116 | NOTCH2 | 15485 | 12498 | -0.093 | 0.1193 | No | ||
| 117 | PAQR6 | 15551 12756 7591 | 13093 | -0.124 | 0.0879 | No | ||
| 118 | TNRC4 | 15514 | 13180 | -0.131 | 0.0841 | No | ||
| 119 | MAN1A2 | 5076 5075 | 13233 | -0.135 | 0.0822 | No | ||
| 120 | THBS3 | 15542 1796 | 13345 | -0.142 | 0.0771 | No | ||
| 121 | LMNA | 4999 15290 1778 9280 | 13455 | -0.151 | 0.0722 | No | ||
| 122 | TIMM23 | 7004 5768 | 13546 | -0.160 | 0.0683 | No | ||
| 123 | PTPRJ | 9664 | 13584 | -0.163 | 0.0674 | No | ||
| 124 | KIRREL3 | 12571 | 13671 | -0.172 | 0.0639 | No | ||
| 125 | ANXA8 | 22046 | 13726 | -0.178 | 0.0621 | No | ||
| 126 | PRKCDBP | 17706 | 13818 | -0.187 | 0.0584 | No | ||
| 127 | HOOK1 | 2395 13418 8102 16173 | 14045 | -0.211 | 0.0475 | No | ||
| 128 | DAB1 | 4594 8834 | 14116 | -0.221 | 0.0451 | No | ||
| 129 | RTN4RL2 | 14544 | 14178 | -0.230 | 0.0433 | No | ||
| 130 | JAM3 | 8197 | 14266 | -0.243 | 0.0402 | No | ||
| 131 | OVOL1 | 23983 | 14503 | -0.283 | 0.0292 | No | ||
| 132 | POGZ | 6031 1911 | 14637 | -0.315 | 0.0241 | No | ||
| 133 | OVGP1 | 4519 8740 | 14773 | -0.350 | 0.0191 | No | ||
| 134 | DEDD | 14056 | 15180 | -0.485 | 0.0002 | No | ||
| 135 | CUL2 | 23630 | 15304 | -0.536 | -0.0030 | No | ||
| 136 | RBM14 | 3673 12087 | 15321 | -0.543 | -0.0003 | No | ||
| 137 | IFRG15 | 7222 | 15463 | -0.636 | -0.0038 | No | ||
| 138 | ADD3 | 23812 3694 | 15544 | -0.676 | -0.0038 | No | ||
| 139 | CHD4 | 8418 17281 4225 | 15754 | -0.826 | -0.0097 | No | ||
| 140 | IPO13 | 15784 | 15881 | -0.920 | -0.0106 | No | ||
| 141 | RAB18 | 1972 5342 | 16023 | -1.008 | -0.0117 | No | ||
| 142 | EHD1 | 23791 | 16354 | -1.280 | -0.0213 | No | ||
| 143 | CACNA1S | 14111 | 16389 | -1.325 | -0.0145 | No | ||
| 144 | HDGF | 9082 4846 4845 | 16438 | -1.371 | -0.0082 | No | ||
| 145 | ST5 | 17682 | 16679 | -1.614 | -0.0107 | No | ||
| 146 | USP5 | 17004 | 17036 | -2.078 | -0.0165 | No | ||
| 147 | PADI2 | 5238 9545 | 17078 | -2.147 | -0.0048 | No | ||
| 148 | SWAP70 | 18126 5553 9948 | 17558 | -3.005 | -0.0113 | No | ||
| 149 | CD2 | 15223 | 18168 | -4.850 | -0.0128 | No | ||
| 150 | MTX1 | 15282 1918 | 18360 | -5.705 | 0.0139 | No |