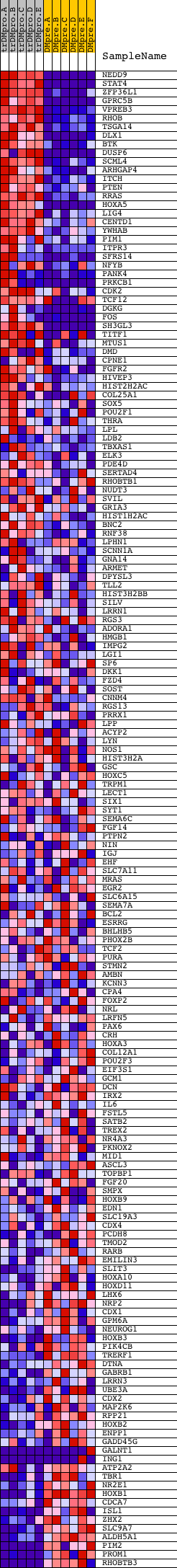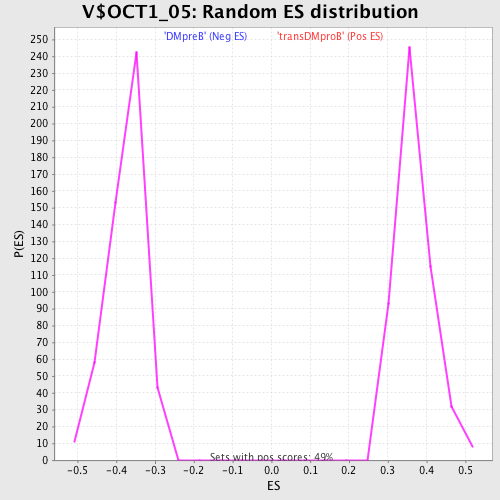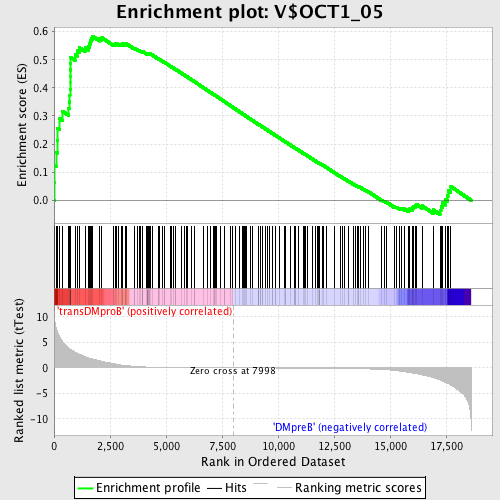
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$OCT1_05 |
| Enrichment Score (ES) | 0.58421487 |
| Normalized Enrichment Score (NES) | 1.5883734 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.051410917 |
| FWER p-Value | 0.362 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NEDD9 | 3206 21483 | 16 | 9.974 | 0.0627 | Yes | ||
| 2 | STAT4 | 14251 9907 | 25 | 9.695 | 0.1241 | Yes | ||
| 3 | ZFP36L1 | 4453 4454 | 88 | 7.945 | 0.1713 | Yes | ||
| 4 | GPRC5B | 17662 | 148 | 7.065 | 0.2132 | Yes | ||
| 5 | VPREB3 | 19984 | 162 | 6.932 | 0.2566 | Yes | ||
| 6 | RHOB | 4411 | 251 | 6.073 | 0.2906 | Yes | ||
| 7 | TSGA14 | 17194 | 364 | 5.178 | 0.3175 | Yes | ||
| 8 | DLX1 | 14988 | 648 | 3.891 | 0.3269 | Yes | ||
| 9 | BTK | 24061 | 669 | 3.794 | 0.3500 | Yes | ||
| 10 | DUSP6 | 19891 3399 | 703 | 3.694 | 0.3718 | Yes | ||
| 11 | SCML4 | 20042 | 710 | 3.673 | 0.3949 | Yes | ||
| 12 | ARHGAP4 | 9343 | 716 | 3.651 | 0.4179 | Yes | ||
| 13 | ITCH | 4928 9185 | 718 | 3.648 | 0.4411 | Yes | ||
| 14 | PTEN | 5305 | 722 | 3.641 | 0.4641 | Yes | ||
| 15 | RRAS | 18256 | 735 | 3.612 | 0.4865 | Yes | ||
| 16 | HOXA5 | 17149 | 746 | 3.576 | 0.5087 | Yes | ||
| 17 | LIG4 | 11454 | 935 | 3.083 | 0.5182 | Yes | ||
| 18 | CENTD1 | 16529 | 1028 | 2.872 | 0.5315 | Yes | ||
| 19 | YWHAB | 14744 | 1122 | 2.677 | 0.5435 | Yes | ||
| 20 | PIM1 | 23308 | 1381 | 2.178 | 0.5434 | Yes | ||
| 21 | ITPR3 | 9195 | 1545 | 1.926 | 0.5469 | Yes | ||
| 22 | SFRS14 | 10622 | 1590 | 1.875 | 0.5564 | Yes | ||
| 23 | NFYB | 5173 | 1607 | 1.850 | 0.5673 | Yes | ||
| 24 | PANK4 | 15972 | 1652 | 1.785 | 0.5763 | Yes | ||
| 25 | PRKCB1 | 1693 9574 | 1709 | 1.712 | 0.5842 | Yes | ||
| 26 | CDK2 | 3438 3373 19592 3322 | 2032 | 1.374 | 0.5755 | No | ||
| 27 | TCF12 | 10044 3125 | 2134 | 1.282 | 0.5782 | No | ||
| 28 | DGKG | 4282 | 2651 | 0.827 | 0.5555 | No | ||
| 29 | FOS | 21202 | 2723 | 0.766 | 0.5565 | No | ||
| 30 | SH3GL3 | 18201 | 2790 | 0.702 | 0.5574 | No | ||
| 31 | TITF1 | 10180 21062 | 2893 | 0.618 | 0.5558 | No | ||
| 32 | MTUS1 | 4162 158 | 2997 | 0.555 | 0.5538 | No | ||
| 33 | DMD | 24295 2647 | 3054 | 0.526 | 0.5541 | No | ||
| 34 | CPNE1 | 11186 | 3061 | 0.519 | 0.5571 | No | ||
| 35 | FGFR2 | 1917 4722 1119 | 3175 | 0.460 | 0.5539 | No | ||
| 36 | HIVEP3 | 4973 | 3184 | 0.456 | 0.5564 | No | ||
| 37 | HIST2H2AC | 6650 | 3210 | 0.447 | 0.5579 | No | ||
| 38 | COL25A1 | 8056 1837 1844 1915 13376 | 3573 | 0.292 | 0.5401 | No | ||
| 39 | SOX5 | 9850 1044 5480 16937 | 3575 | 0.291 | 0.5419 | No | ||
| 40 | POU2F1 | 5275 3989 4065 4010 | 3742 | 0.245 | 0.5345 | No | ||
| 41 | THRA | 1447 10171 1406 | 3826 | 0.224 | 0.5314 | No | ||
| 42 | LPL | 9283 5003 | 3862 | 0.216 | 0.5309 | No | ||
| 43 | LDB2 | 16538 4989 | 3934 | 0.196 | 0.5283 | No | ||
| 44 | TBXAS1 | 17480 1105 | 3952 | 0.191 | 0.5286 | No | ||
| 45 | ELK3 | 4663 94 3388 | 3955 | 0.190 | 0.5297 | No | ||
| 46 | PDE4D | 10722 6235 | 4135 | 0.158 | 0.5210 | No | ||
| 47 | SERTAD4 | 13715 | 4149 | 0.155 | 0.5212 | No | ||
| 48 | RHOBTB1 | 12789 | 4176 | 0.151 | 0.5208 | No | ||
| 49 | NUDT3 | 23056 | 4180 | 0.150 | 0.5216 | No | ||
| 50 | SVIL | 1990 1942 10334 1976 23626 1946 1945 2041 | 4185 | 0.150 | 0.5223 | No | ||
| 51 | GRIA3 | 24349 11996 2649 | 4231 | 0.143 | 0.5208 | No | ||
| 52 | HIST1H2AC | 6648 | 4232 | 0.143 | 0.5217 | No | ||
| 53 | BNC2 | 15850 | 4247 | 0.142 | 0.5219 | No | ||
| 54 | RNF38 | 7862 2328 13115 | 4254 | 0.140 | 0.5224 | No | ||
| 55 | LPHN1 | 6850 174 | 4315 | 0.133 | 0.5200 | No | ||
| 56 | SCNN1A | 17278 | 4370 | 0.123 | 0.5179 | No | ||
| 57 | GNA14 | 23907 | 4649 | 0.091 | 0.5034 | No | ||
| 58 | ARMET | 19005 | 4660 | 0.090 | 0.5034 | No | ||
| 59 | DPYSL3 | 10252 | 4712 | 0.086 | 0.5012 | No | ||
| 60 | TLL2 | 23680 | 4858 | 0.075 | 0.4938 | No | ||
| 61 | HIST3H2BB | 11796 | 4923 | 0.072 | 0.4908 | No | ||
| 62 | SILV | 16130 | 4932 | 0.072 | 0.4908 | No | ||
| 63 | LRRN1 | 17343 | 5209 | 0.058 | 0.4762 | No | ||
| 64 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 5251 | 0.056 | 0.4744 | No | ||
| 65 | ADORA1 | 13831 | 5319 | 0.053 | 0.4711 | No | ||
| 66 | HMGB1 | 9094 4855 | 5397 | 0.050 | 0.4672 | No | ||
| 67 | IMPG2 | 22731 | 5427 | 0.049 | 0.4659 | No | ||
| 68 | LGI1 | 23867 | 5675 | 0.041 | 0.4528 | No | ||
| 69 | SP6 | 8178 13678 13679 | 5835 | 0.037 | 0.4444 | No | ||
| 70 | DKK1 | 23700 | 5901 | 0.035 | 0.4411 | No | ||
| 71 | FZD4 | 18193 | 5950 | 0.034 | 0.4387 | No | ||
| 72 | SOST | 20210 | 6128 | 0.030 | 0.4293 | No | ||
| 73 | CNNM4 | 14273 | 6256 | 0.027 | 0.4226 | No | ||
| 74 | RGS13 | 13811 | 6282 | 0.027 | 0.4214 | No | ||
| 75 | PRRX1 | 9595 5271 | 6683 | 0.019 | 0.3999 | No | ||
| 76 | LPP | 5570 | 6854 | 0.016 | 0.3908 | No | ||
| 77 | ACYP2 | 20508 | 7000 | 0.014 | 0.3830 | No | ||
| 78 | LYN | 16281 | 7117 | 0.012 | 0.3768 | No | ||
| 79 | NOS1 | 9474 | 7156 | 0.012 | 0.3748 | No | ||
| 80 | HIST3H2A | 6647 | 7174 | 0.011 | 0.3739 | No | ||
| 81 | GSC | 20990 | 7204 | 0.011 | 0.3724 | No | ||
| 82 | HOXC5 | 22340 | 7250 | 0.010 | 0.3701 | No | ||
| 83 | TRPM1 | 10846 474 9395 494 | 7430 | 0.007 | 0.3604 | No | ||
| 84 | LECT1 | 21740 | 7605 | 0.005 | 0.3510 | No | ||
| 85 | SIX1 | 9821 | 7626 | 0.005 | 0.3500 | No | ||
| 86 | SYT1 | 5565 | 7856 | 0.002 | 0.3375 | No | ||
| 87 | SEMA6C | 1797 5424 | 7952 | 0.001 | 0.3324 | No | ||
| 88 | FGF14 | 21716 3005 3463 | 8086 | -0.001 | 0.3252 | No | ||
| 89 | PTPN2 | 23414 | 8291 | -0.004 | 0.3142 | No | ||
| 90 | NIN | 9463 2103 | 8421 | -0.006 | 0.3072 | No | ||
| 91 | IGJ | 4903 | 8463 | -0.006 | 0.3050 | No | ||
| 92 | EHF | 4657 | 8491 | -0.007 | 0.3036 | No | ||
| 93 | SLC7A11 | 6473 | 8561 | -0.007 | 0.2999 | No | ||
| 94 | MRAS | 9409 | 8608 | -0.008 | 0.2975 | No | ||
| 95 | EGR2 | 8886 | 8783 | -0.011 | 0.2881 | No | ||
| 96 | SLC6A15 | 8335 3321 | 8861 | -0.012 | 0.2840 | No | ||
| 97 | SEMA7A | 19426 9803 | 9104 | -0.015 | 0.2710 | No | ||
| 98 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 9115 | -0.015 | 0.2705 | No | ||
| 99 | ESRRG | 6447 | 9116 | -0.015 | 0.2706 | No | ||
| 100 | BHLHB5 | 15629 | 9205 | -0.017 | 0.2660 | No | ||
| 101 | PHOX2B | 16524 | 9212 | -0.017 | 0.2657 | No | ||
| 102 | TCF2 | 1395 20727 | 9294 | -0.018 | 0.2615 | No | ||
| 103 | PURA | 9670 | 9307 | -0.018 | 0.2609 | No | ||
| 104 | STMN2 | 15638 | 9422 | -0.020 | 0.2549 | No | ||
| 105 | AMBN | 4380 | 9530 | -0.021 | 0.2492 | No | ||
| 106 | KCNN3 | 15538 1913 | 9608 | -0.022 | 0.2452 | No | ||
| 107 | CPA4 | 7757 | 9733 | -0.024 | 0.2386 | No | ||
| 108 | FOXP2 | 4312 17523 8523 | 9742 | -0.024 | 0.2383 | No | ||
| 109 | NRL | 9484 | 9863 | -0.026 | 0.2320 | No | ||
| 110 | LRFN5 | 6213 | 9892 | -0.027 | 0.2306 | No | ||
| 111 | PAX6 | 5223 | 10064 | -0.030 | 0.2216 | No | ||
| 112 | CRH | 8784 | 10293 | -0.034 | 0.2094 | No | ||
| 113 | HOXA3 | 9108 1075 | 10322 | -0.034 | 0.2081 | No | ||
| 114 | COL12A1 | 3143 19054 | 10329 | -0.034 | 0.2080 | No | ||
| 115 | POU2F3 | 9602 | 10537 | -0.038 | 0.1970 | No | ||
| 116 | EIF3S1 | 905 8114 | 10712 | -0.041 | 0.1878 | No | ||
| 117 | GCM1 | 19372 | 10717 | -0.041 | 0.1879 | No | ||
| 118 | DCN | 19897 4601 8840 3366 | 10772 | -0.042 | 0.1852 | No | ||
| 119 | IRX2 | 4927 | 10775 | -0.042 | 0.1854 | No | ||
| 120 | IL6 | 16895 | 10906 | -0.045 | 0.1786 | No | ||
| 121 | FSTL5 | 10018 | 11123 | -0.049 | 0.1672 | No | ||
| 122 | SATB2 | 5604 | 11162 | -0.050 | 0.1655 | No | ||
| 123 | TREX2 | 24137 | 11205 | -0.051 | 0.1635 | No | ||
| 124 | NR4A3 | 9473 16212 5183 | 11305 | -0.053 | 0.1585 | No | ||
| 125 | PKNOX2 | 3090 5511 9889 | 11519 | -0.058 | 0.1473 | No | ||
| 126 | MID1 | 5097 5098 | 11671 | -0.062 | 0.1395 | No | ||
| 127 | ASCL3 | 17681 | 11761 | -0.064 | 0.1351 | No | ||
| 128 | TOPBP1 | 10659 | 11806 | -0.066 | 0.1331 | No | ||
| 129 | FGF20 | 18892 | 11835 | -0.067 | 0.1320 | No | ||
| 130 | SMPX | 24220 | 11845 | -0.067 | 0.1320 | No | ||
| 131 | HOXB9 | 20689 | 11854 | -0.067 | 0.1320 | No | ||
| 132 | EDN1 | 21658 | 11974 | -0.071 | 0.1260 | No | ||
| 133 | SLC19A3 | 8152 | 12036 | -0.073 | 0.1231 | No | ||
| 134 | CDX4 | 24273 | 12165 | -0.078 | 0.1167 | No | ||
| 135 | PCDH8 | 21739 3017 | 12521 | -0.094 | 0.0981 | No | ||
| 136 | TMOD2 | 6946 | 12786 | -0.107 | 0.0844 | No | ||
| 137 | RARB | 3011 21915 | 12871 | -0.111 | 0.0806 | No | ||
| 138 | EMILIN3 | 11376 | 12958 | -0.116 | 0.0767 | No | ||
| 139 | SLIT3 | 20925 | 13152 | -0.129 | 0.0670 | No | ||
| 140 | HOXA10 | 17145 989 | 13155 | -0.129 | 0.0677 | No | ||
| 141 | HOXD11 | 9112 | 13351 | -0.142 | 0.0581 | No | ||
| 142 | LHX6 | 9275 2914 | 13474 | -0.153 | 0.0524 | No | ||
| 143 | NRP2 | 5194 9486 5195 | 13529 | -0.158 | 0.0505 | No | ||
| 144 | CDX1 | 23429 | 13604 | -0.165 | 0.0475 | No | ||
| 145 | GPM6A | 3834 10618 | 13606 | -0.166 | 0.0485 | No | ||
| 146 | NEUROG1 | 21446 | 13662 | -0.171 | 0.0466 | No | ||
| 147 | HOXB3 | 20685 | 13829 | -0.188 | 0.0388 | No | ||
| 148 | PIK4CB | 1875 15511 | 13914 | -0.196 | 0.0355 | No | ||
| 149 | TRERF1 | 1546 23213 | 14037 | -0.211 | 0.0303 | No | ||
| 150 | DTNA | 8870 4647 23611 1991 | 14636 | -0.315 | -0.0001 | No | ||
| 151 | GABRB1 | 16832 | 14734 | -0.341 | -0.0032 | No | ||
| 152 | LRRN3 | 21077 | 14836 | -0.366 | -0.0064 | No | ||
| 153 | UBE3A | 1209 1084 1830 | 15203 | -0.494 | -0.0231 | No | ||
| 154 | CDX2 | 16289 | 15282 | -0.527 | -0.0239 | No | ||
| 155 | MAP2K6 | 20614 1414 | 15424 | -0.607 | -0.0277 | No | ||
| 156 | RPP21 | 22993 | 15519 | -0.663 | -0.0286 | No | ||
| 157 | HOXB2 | 8346 | 15644 | -0.737 | -0.0306 | No | ||
| 158 | ENPP1 | 19804 | 15834 | -0.885 | -0.0352 | No | ||
| 159 | GADD45G | 21637 | 15858 | -0.901 | -0.0307 | No | ||
| 160 | GALNT1 | 8998 23609 8999 4750 | 15991 | -0.999 | -0.0315 | No | ||
| 161 | ING1 | 6440 6439 | 15992 | -0.999 | -0.0252 | No | ||
| 162 | ATP2A2 | 4421 3481 | 16060 | -1.027 | -0.0223 | No | ||
| 163 | TBR1 | 83 | 16147 | -1.087 | -0.0200 | No | ||
| 164 | NR2E1 | 19780 | 16161 | -1.099 | -0.0137 | No | ||
| 165 | HOXB1 | 20684 | 16425 | -1.354 | -0.0193 | No | ||
| 166 | CDCA7 | 12437 | 16925 | -1.927 | -0.0341 | No | ||
| 167 | ISL1 | 21338 | 17228 | -2.358 | -0.0354 | No | ||
| 168 | ZHX2 | 11842 | 17273 | -2.466 | -0.0221 | No | ||
| 169 | SLC9A7 | 24177 | 17326 | -2.578 | -0.0085 | No | ||
| 170 | ALDH5A1 | 21511 | 17481 | -2.867 | 0.0014 | No | ||
| 171 | PIM2 | 5249 9564 9565 | 17560 | -3.008 | 0.0164 | No | ||
| 172 | PROM1 | 5293 | 17585 | -3.048 | 0.0345 | No | ||
| 173 | RHOBTB3 | 7849 | 17713 | -3.351 | 0.0490 | No |