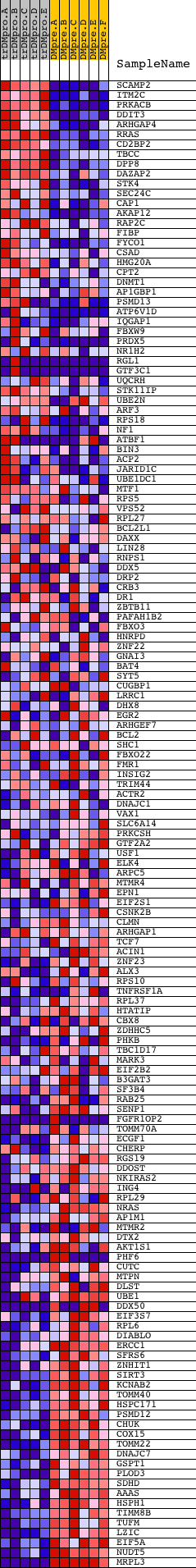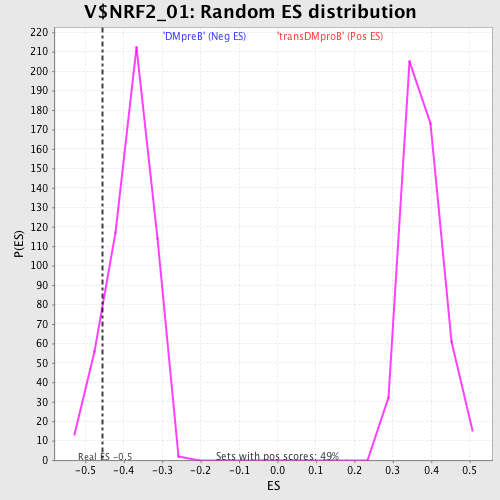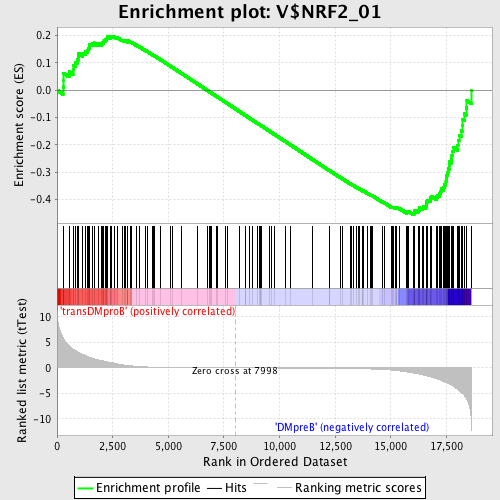
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | V$NRF2_01 |
| Enrichment Score (ES) | -0.45569852 |
| Normalized Enrichment Score (NES) | -1.1897091 |
| Nominal p-value | 0.105058365 |
| FDR q-value | 0.7436318 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SCAMP2 | 19430 | 267 | 5.895 | 0.0120 | No | ||
| 2 | ITM2C | 14204 | 282 | 5.751 | 0.0371 | No | ||
| 3 | PRKACB | 15140 | 286 | 5.707 | 0.0626 | No | ||
| 4 | DDIT3 | 19857 | 544 | 4.335 | 0.0681 | No | ||
| 5 | ARHGAP4 | 9343 | 716 | 3.651 | 0.0752 | No | ||
| 6 | RRAS | 18256 | 735 | 3.612 | 0.0905 | No | ||
| 7 | CD2BP2 | 17621 | 814 | 3.387 | 0.1015 | No | ||
| 8 | TBCC | 23215 | 897 | 3.189 | 0.1113 | No | ||
| 9 | DPP8 | 19408 | 943 | 3.063 | 0.1227 | No | ||
| 10 | DAZAP2 | 10758 | 967 | 3.004 | 0.1349 | No | ||
| 11 | STK4 | 14743 | 1158 | 2.598 | 0.1363 | No | ||
| 12 | SEC24C | 22086 | 1269 | 2.375 | 0.1410 | No | ||
| 13 | CAP1 | 8684 | 1350 | 2.228 | 0.1467 | No | ||
| 14 | AKAP12 | 17600 | 1392 | 2.167 | 0.1542 | No | ||
| 15 | RAP2C | 24156 | 1439 | 2.082 | 0.1610 | No | ||
| 16 | FIBP | 7195 | 1469 | 2.036 | 0.1686 | No | ||
| 17 | FYCO1 | 18952 | 1583 | 1.878 | 0.1709 | No | ||
| 18 | CSAD | 10899 | 1685 | 1.740 | 0.1733 | No | ||
| 19 | HMG20A | 19436 | 1864 | 1.552 | 0.1706 | No | ||
| 20 | CPT2 | 15812 900 | 1990 | 1.412 | 0.1702 | No | ||
| 21 | DNMT1 | 19217 | 2021 | 1.379 | 0.1747 | No | ||
| 22 | AP1GBP1 | 20725 | 2098 | 1.313 | 0.1765 | No | ||
| 23 | PSMD13 | 18019 3897 | 2106 | 1.304 | 0.1820 | No | ||
| 24 | ATP6V1D | 7872 | 2157 | 1.261 | 0.1850 | No | ||
| 25 | IQGAP1 | 6619 | 2207 | 1.211 | 0.1877 | No | ||
| 26 | FBXW9 | 18540 | 2239 | 1.172 | 0.1913 | No | ||
| 27 | PRDX5 | 7053 3732 | 2242 | 1.172 | 0.1965 | No | ||
| 28 | NR1H2 | 17846 | 2387 | 1.040 | 0.1934 | No | ||
| 29 | RGL1 | 9723 4072 | 2398 | 1.033 | 0.1974 | No | ||
| 30 | GTF3C1 | 10607 17646 | 2465 | 0.997 | 0.1983 | No | ||
| 31 | UQCRH | 12378 | 2594 | 0.878 | 0.1954 | No | ||
| 32 | STK11IP | 14212 | 2708 | 0.778 | 0.1927 | No | ||
| 33 | UBE2N | 8216 19898 | 2919 | 0.603 | 0.1841 | No | ||
| 34 | ARF3 | 22137 | 3019 | 0.547 | 0.1812 | No | ||
| 35 | RPS18 | 1529 5397 1603 | 3093 | 0.504 | 0.1795 | No | ||
| 36 | NF1 | 5165 | 3151 | 0.469 | 0.1785 | No | ||
| 37 | ATBF1 | 916 8633 | 3161 | 0.464 | 0.1801 | No | ||
| 38 | BIN3 | 21969 | 3173 | 0.461 | 0.1816 | No | ||
| 39 | ACP2 | 4318 | 3295 | 0.405 | 0.1768 | No | ||
| 40 | JARID1C | 5460 2563 | 3353 | 0.378 | 0.1755 | No | ||
| 41 | UBE1DC1 | 19020 | 3555 | 0.299 | 0.1659 | No | ||
| 42 | MTF1 | 5128 9421 | 3694 | 0.257 | 0.1596 | No | ||
| 43 | RPS5 | 18391 | 3988 | 0.184 | 0.1446 | No | ||
| 44 | VPS52 | 23287 1622 | 4069 | 0.169 | 0.1410 | No | ||
| 45 | RPL27 | 9740 | 4270 | 0.139 | 0.1308 | No | ||
| 46 | BCL2L1 | 4440 2930 8652 | 4316 | 0.132 | 0.1289 | No | ||
| 47 | DAXX | 23290 | 4398 | 0.119 | 0.1251 | No | ||
| 48 | LIN28 | 15723 | 4666 | 0.090 | 0.1110 | No | ||
| 49 | RNPS1 | 9730 23361 | 5114 | 0.062 | 0.0871 | No | ||
| 50 | DDX5 | 4606 8843 4607 | 5187 | 0.059 | 0.0835 | No | ||
| 51 | DRP2 | 24254 | 5583 | 0.044 | 0.0623 | No | ||
| 52 | CRB3 | 23176 | 6325 | 0.026 | 0.0223 | No | ||
| 53 | DR1 | 16762 | 6756 | 0.018 | -0.0010 | No | ||
| 54 | ZBTB11 | 11309 | 6853 | 0.016 | -0.0061 | No | ||
| 55 | PAFAH1B2 | 5221 9525 | 6892 | 0.015 | -0.0081 | No | ||
| 56 | FBXO3 | 2662 14934 | 6937 | 0.015 | -0.0104 | No | ||
| 57 | HNRPD | 8645 | 7181 | 0.011 | -0.0235 | No | ||
| 58 | ZNF22 | 12497 7417 | 7214 | 0.011 | -0.0252 | No | ||
| 59 | GNAI3 | 15198 | 7576 | 0.005 | -0.0447 | No | ||
| 60 | BAT4 | 8172 8173 | 7671 | 0.004 | -0.0498 | No | ||
| 61 | SYT5 | 17986 | 8182 | -0.003 | -0.0774 | No | ||
| 62 | CUGBP1 | 2805 8819 4576 2924 | 8484 | -0.007 | -0.0937 | No | ||
| 63 | LRRC1 | 19059 3129 | 8489 | -0.007 | -0.0938 | No | ||
| 64 | DHX8 | 20649 | 8662 | -0.009 | -0.1031 | No | ||
| 65 | EGR2 | 8886 | 8783 | -0.011 | -0.1096 | No | ||
| 66 | ARHGEF7 | 3823 | 8994 | -0.014 | -0.1209 | No | ||
| 67 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 9115 | -0.015 | -0.1273 | No | ||
| 68 | SHC1 | 9813 9812 5430 | 9137 | -0.016 | -0.1284 | No | ||
| 69 | FBXO22 | 19442 | 9141 | -0.016 | -0.1285 | No | ||
| 70 | FMR1 | 24321 | 9187 | -0.016 | -0.1308 | No | ||
| 71 | INSIG2 | 3944 4124 4073 | 9534 | -0.021 | -0.1495 | No | ||
| 72 | TRIM44 | 8164 13487 | 9617 | -0.023 | -0.1538 | No | ||
| 73 | ACTR2 | 20523 | 9775 | -0.025 | -0.1622 | No | ||
| 74 | DNAJC1 | 8854 | 10282 | -0.033 | -0.1894 | No | ||
| 75 | VAX1 | 23636 | 10511 | -0.037 | -0.2016 | No | ||
| 76 | SLC6A14 | 12168 | 11467 | -0.056 | -0.2531 | No | ||
| 77 | PRKCSH | 19535 | 12236 | -0.081 | -0.2943 | No | ||
| 78 | GTF2A2 | 10654 | 12265 | -0.082 | -0.2955 | No | ||
| 79 | USF1 | 5832 10260 10261 | 12736 | -0.104 | -0.3205 | No | ||
| 80 | ELK4 | 8895 4664 3937 | 12844 | -0.110 | -0.3258 | No | ||
| 81 | ARPC5 | 12580 7487 | 13172 | -0.130 | -0.3429 | No | ||
| 82 | MTMR4 | 20712 | 13235 | -0.135 | -0.3456 | No | ||
| 83 | EPN1 | 18401 | 13314 | -0.140 | -0.3492 | No | ||
| 84 | EIF2S1 | 4658 | 13442 | -0.150 | -0.3554 | No | ||
| 85 | CSNK2B | 23008 | 13539 | -0.159 | -0.3599 | No | ||
| 86 | CLMN | 20988 2119 2104 | 13607 | -0.166 | -0.3628 | No | ||
| 87 | ARHGAP1 | 6001 10448 | 13740 | -0.179 | -0.3691 | No | ||
| 88 | TCF7 | 1467 20466 | 13747 | -0.180 | -0.3687 | No | ||
| 89 | ACIN1 | 3319 2807 21832 | 13776 | -0.182 | -0.3694 | No | ||
| 90 | ZNF23 | 6149 | 13936 | -0.199 | -0.3771 | No | ||
| 91 | ALX3 | 15456 | 14101 | -0.219 | -0.3850 | No | ||
| 92 | RPS10 | 12464 | 14133 | -0.223 | -0.3856 | No | ||
| 93 | TNFRSF1A | 1181 10206 | 14170 | -0.228 | -0.3866 | No | ||
| 94 | RPL37 | 12502 7421 22521 | 14614 | -0.310 | -0.4092 | No | ||
| 95 | HTATIP | 3690 | 14713 | -0.337 | -0.4130 | No | ||
| 96 | CBX8 | 11401 | 15033 | -0.425 | -0.4283 | No | ||
| 97 | ZDHHC5 | 14547 | 15079 | -0.445 | -0.4288 | No | ||
| 98 | PHKB | 18537 3873 | 15115 | -0.460 | -0.4286 | No | ||
| 99 | TBC1D17 | 6121 | 15220 | -0.501 | -0.4320 | No | ||
| 100 | MARK3 | 9369 | 15234 | -0.507 | -0.4304 | No | ||
| 101 | EIF2B2 | 21204 | 15254 | -0.516 | -0.4291 | No | ||
| 102 | B3GAT3 | 23938 | 15383 | -0.578 | -0.4334 | No | ||
| 103 | SF3B4 | 22269 | 15726 | -0.807 | -0.4483 | No | ||
| 104 | RAB25 | 15287 | 15734 | -0.810 | -0.4451 | No | ||
| 105 | SENP1 | 10301 | 15780 | -0.844 | -0.4437 | No | ||
| 106 | FGFR1OP2 | 12544 12543 7457 | 16002 | -1.001 | -0.4512 | Yes | ||
| 107 | TOMM70A | 6608 | 16056 | -1.025 | -0.4495 | Yes | ||
| 108 | ECGF1 | 22160 | 16080 | -1.039 | -0.4460 | Yes | ||
| 109 | CHERP | 11367 | 16081 | -1.040 | -0.4414 | Yes | ||
| 110 | RGS19 | 14303 | 16222 | -1.156 | -0.4438 | Yes | ||
| 111 | DDOST | 16021 | 16255 | -1.186 | -0.4402 | Yes | ||
| 112 | NKIRAS2 | 12979 | 16291 | -1.219 | -0.4366 | Yes | ||
| 113 | ING4 | 6602 1155 1102 11371 | 16292 | -1.220 | -0.4311 | Yes | ||
| 114 | RPL29 | 9741 | 16435 | -1.368 | -0.4327 | Yes | ||
| 115 | NRAS | 5191 | 16449 | -1.395 | -0.4271 | Yes | ||
| 116 | AP1M1 | 18571 | 16582 | -1.540 | -0.4273 | Yes | ||
| 117 | MTMR2 | 8069 3003 | 16590 | -1.544 | -0.4208 | Yes | ||
| 118 | DTX2 | 16674 3488 | 16595 | -1.548 | -0.4140 | Yes | ||
| 119 | AKT1S1 | 12557 | 16605 | -1.556 | -0.4075 | Yes | ||
| 120 | PHF6 | 2648 24340 | 16655 | -1.589 | -0.4031 | Yes | ||
| 121 | CUTC | 3711 | 16775 | -1.711 | -0.4018 | Yes | ||
| 122 | MTPN | 17183 | 16779 | -1.716 | -0.3943 | Yes | ||
| 123 | DLST | 8131 | 16811 | -1.766 | -0.3880 | Yes | ||
| 124 | UBE1 | 24370 2551 | 17042 | -2.087 | -0.3911 | Yes | ||
| 125 | DDX50 | 3324 19749 | 17112 | -2.188 | -0.3850 | Yes | ||
| 126 | EIF3S7 | 22226 | 17177 | -2.255 | -0.3784 | Yes | ||
| 127 | RPL6 | 9747 | 17219 | -2.333 | -0.3701 | Yes | ||
| 128 | DIABLO | 16375 | 17272 | -2.465 | -0.3618 | Yes | ||
| 129 | ERCC1 | 1235 1045 1646 | 17375 | -2.658 | -0.3554 | Yes | ||
| 130 | SFRS6 | 14751 | 17415 | -2.724 | -0.3453 | Yes | ||
| 131 | ZNHIT1 | 16334 | 17478 | -2.863 | -0.3358 | Yes | ||
| 132 | SIRT3 | 17569 3713 | 17486 | -2.875 | -0.3233 | Yes | ||
| 133 | KCNAB2 | 15655 | 17520 | -2.947 | -0.3118 | Yes | ||
| 134 | TOMM40 | 6997 | 17562 | -3.010 | -0.3005 | Yes | ||
| 135 | HSPC171 | 18494 | 17574 | -3.032 | -0.2875 | Yes | ||
| 136 | PSMD12 | 20621 | 17621 | -3.111 | -0.2760 | Yes | ||
| 137 | CHUK | 23665 | 17633 | -3.139 | -0.2625 | Yes | ||
| 138 | COX15 | 5931 | 17718 | -3.365 | -0.2520 | Yes | ||
| 139 | TOMM22 | 5863 | 17744 | -3.415 | -0.2380 | Yes | ||
| 140 | DNAJC7 | 20227 | 17784 | -3.518 | -0.2243 | Yes | ||
| 141 | GSPT1 | 9046 | 17814 | -3.603 | -0.2097 | Yes | ||
| 142 | PLOD3 | 16666 | 18006 | -4.223 | -0.2011 | Yes | ||
| 143 | SDHD | 19125 | 18022 | -4.265 | -0.1827 | Yes | ||
| 144 | AAAS | 22111 2274 | 18068 | -4.474 | -0.1651 | Yes | ||
| 145 | HSPH1 | 16282 | 18166 | -4.828 | -0.1486 | Yes | ||
| 146 | TIMM8B | 11391 | 18205 | -4.967 | -0.1284 | Yes | ||
| 147 | TUFM | 10608 | 18239 | -5.070 | -0.1074 | Yes | ||
| 148 | LZIC | 15988 | 18315 | -5.457 | -0.0870 | Yes | ||
| 149 | EIF5A | 11345 20379 6590 | 18392 | -5.951 | -0.0643 | Yes | ||
| 150 | NUDT5 | 15121 | 18421 | -6.118 | -0.0384 | Yes | ||
| 151 | MRPL3 | 3116 19335 | 18611 | -10.888 | 0.0003 | Yes |