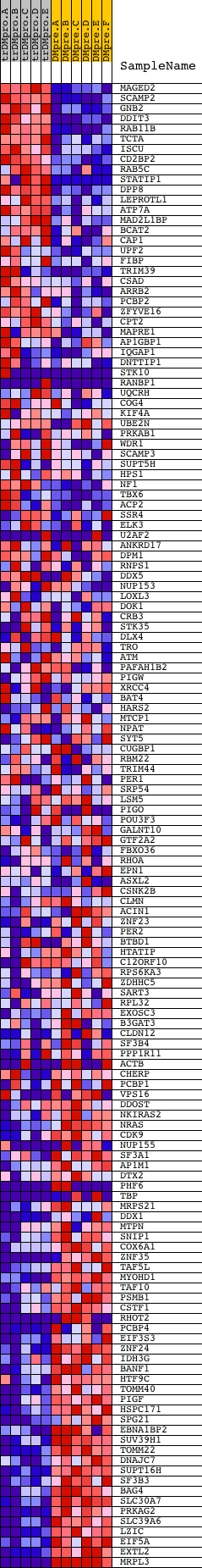
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
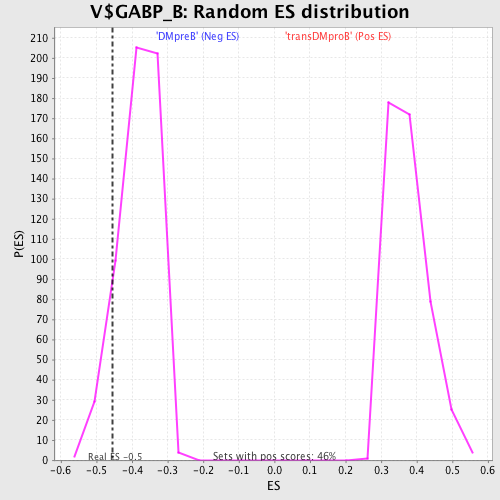
| GeneSet | V$GABP_B |
| Enrichment Score (ES) | -0.45346606 |
| Normalized Enrichment Score (NES) | -1.1924483 |
| Nominal p-value | 0.0961183 |
| FDR q-value | 0.76453143 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MAGED2 | 24034 | 207 | 6.431 | 0.0163 | No | ||
| 2 | SCAMP2 | 19430 | 267 | 5.895 | 0.0382 | No | ||
| 3 | GNB2 | 9026 | 487 | 4.520 | 0.0457 | No | ||
| 4 | DDIT3 | 19857 | 544 | 4.335 | 0.0612 | No | ||
| 5 | RAB11B | 9676 | 558 | 4.276 | 0.0787 | No | ||
| 6 | TCTA | 4167 | 619 | 4.015 | 0.0926 | No | ||
| 7 | ISCU | 12340 | 632 | 3.960 | 0.1089 | No | ||
| 8 | CD2BP2 | 17621 | 814 | 3.387 | 0.1136 | No | ||
| 9 | RAB5C | 20226 | 892 | 3.201 | 0.1231 | No | ||
| 10 | STATIP1 | 12215 2015 | 931 | 3.095 | 0.1342 | No | ||
| 11 | DPP8 | 19408 | 943 | 3.063 | 0.1467 | No | ||
| 12 | LEPROTL1 | 18896 | 1003 | 2.934 | 0.1561 | No | ||
| 13 | ATP7A | 4425 | 1239 | 2.443 | 0.1538 | No | ||
| 14 | MAD2L1BP | 22967 8027 | 1295 | 2.332 | 0.1608 | No | ||
| 15 | BCAT2 | 18244 | 1306 | 2.314 | 0.1701 | No | ||
| 16 | CAP1 | 8684 | 1350 | 2.228 | 0.1773 | No | ||
| 17 | UPF2 | 11591 | 1449 | 2.064 | 0.1808 | No | ||
| 18 | FIBP | 7195 | 1469 | 2.036 | 0.1885 | No | ||
| 19 | TRIM39 | 22994 | 1585 | 1.878 | 0.1903 | No | ||
| 20 | CSAD | 10899 | 1685 | 1.740 | 0.1923 | No | ||
| 21 | ARRB2 | 20806 | 1715 | 1.707 | 0.1981 | No | ||
| 22 | PCBP2 | 9533 | 1814 | 1.620 | 0.1997 | No | ||
| 23 | ZFYVE16 | 21395 | 1843 | 1.576 | 0.2049 | No | ||
| 24 | CPT2 | 15812 900 | 1990 | 1.412 | 0.2030 | No | ||
| 25 | MAPRE1 | 4652 | 2057 | 1.347 | 0.2052 | No | ||
| 26 | AP1GBP1 | 20725 | 2098 | 1.313 | 0.2087 | No | ||
| 27 | IQGAP1 | 6619 | 2207 | 1.211 | 0.2080 | No | ||
| 28 | DNTTIP1 | 14736 | 2262 | 1.150 | 0.2100 | No | ||
| 29 | STK10 | 5532 9916 | 2421 | 1.013 | 0.2057 | No | ||
| 30 | RANBP1 | 9692 5357 | 2445 | 1.000 | 0.2088 | No | ||
| 31 | UQCRH | 12378 | 2594 | 0.878 | 0.2045 | No | ||
| 32 | COG4 | 18465 | 2876 | 0.631 | 0.1920 | No | ||
| 33 | KIF4A | 9221 24281 | 2879 | 0.630 | 0.1946 | No | ||
| 34 | UBE2N | 8216 19898 | 2919 | 0.603 | 0.1950 | No | ||
| 35 | PRKAB1 | 16408 | 2954 | 0.580 | 0.1957 | No | ||
| 36 | WDR1 | 16544 3592 | 3016 | 0.547 | 0.1947 | No | ||
| 37 | SCAMP3 | 15543 | 3075 | 0.514 | 0.1938 | No | ||
| 38 | SUPT5H | 9939 | 3083 | 0.511 | 0.1956 | No | ||
| 39 | HPS1 | 23671 | 3086 | 0.508 | 0.1976 | No | ||
| 40 | NF1 | 5165 | 3151 | 0.469 | 0.1962 | No | ||
| 41 | TBX6 | 1993 18075 | 3155 | 0.467 | 0.1980 | No | ||
| 42 | ACP2 | 4318 | 3295 | 0.405 | 0.1922 | No | ||
| 43 | SSR4 | 24303 5519 | 3589 | 0.287 | 0.1776 | No | ||
| 44 | ELK3 | 4663 94 3388 | 3955 | 0.190 | 0.1586 | No | ||
| 45 | U2AF2 | 5813 5812 5814 | 4319 | 0.132 | 0.1395 | No | ||
| 46 | ANKRD17 | 13496 3474 8170 | 4736 | 0.084 | 0.1174 | No | ||
| 47 | DPM1 | 4636 2882 8860 | 4775 | 0.081 | 0.1157 | No | ||
| 48 | RNPS1 | 9730 23361 | 5114 | 0.062 | 0.0976 | No | ||
| 49 | DDX5 | 4606 8843 4607 | 5187 | 0.059 | 0.0940 | No | ||
| 50 | NUP153 | 21474 | 5681 | 0.041 | 0.0675 | No | ||
| 51 | LOXL3 | 17401 | 5749 | 0.039 | 0.0640 | No | ||
| 52 | DOK1 | 17104 1018 1177 | 5992 | 0.033 | 0.0510 | No | ||
| 53 | CRB3 | 23176 | 6325 | 0.026 | 0.0332 | No | ||
| 54 | STK35 | 14851 | 6327 | 0.026 | 0.0332 | No | ||
| 55 | DLX4 | 20282 1225 | 6380 | 0.025 | 0.0305 | No | ||
| 56 | TRO | 7071 | 6527 | 0.022 | 0.0227 | No | ||
| 57 | ATM | 2976 19115 | 6719 | 0.018 | 0.0124 | No | ||
| 58 | PAFAH1B2 | 5221 9525 | 6892 | 0.015 | 0.0032 | No | ||
| 59 | PIGW | 20312 | 7419 | 0.008 | -0.0252 | No | ||
| 60 | XRCC4 | 4238 8432 | 7532 | 0.006 | -0.0313 | No | ||
| 61 | BAT4 | 8172 8173 | 7671 | 0.004 | -0.0387 | No | ||
| 62 | HARS2 | 23592 14820 | 7675 | 0.004 | -0.0389 | No | ||
| 63 | MTCP1 | 24122 653 | 7918 | 0.001 | -0.0520 | No | ||
| 64 | NPAT | 19452 6356 | 8038 | -0.000 | -0.0584 | No | ||
| 65 | SYT5 | 17986 | 8182 | -0.003 | -0.0661 | No | ||
| 66 | CUGBP1 | 2805 8819 4576 2924 | 8484 | -0.007 | -0.0824 | No | ||
| 67 | RBM22 | 23542 | 9160 | -0.016 | -0.1189 | No | ||
| 68 | TRIM44 | 8164 13487 | 9617 | -0.023 | -0.1435 | No | ||
| 69 | PER1 | 20827 | 9750 | -0.024 | -0.1505 | No | ||
| 70 | SRP54 | 6282 | 10085 | -0.030 | -0.1685 | No | ||
| 71 | LSM5 | 7286 | 10421 | -0.036 | -0.1865 | No | ||
| 72 | PIGO | 15903 2488 2344 | 11004 | -0.046 | -0.2178 | No | ||
| 73 | POU3F3 | 14261 | 11726 | -0.063 | -0.2565 | No | ||
| 74 | GALNT10 | 9345 1269 | 12039 | -0.074 | -0.2731 | No | ||
| 75 | GTF2A2 | 10654 | 12265 | -0.082 | -0.2849 | No | ||
| 76 | FBXO36 | 14205 | 12355 | -0.087 | -0.2894 | No | ||
| 77 | RHOA | 8624 4409 4410 | 13287 | -0.138 | -0.3392 | No | ||
| 78 | EPN1 | 18401 | 13314 | -0.140 | -0.3400 | No | ||
| 79 | ASXL2 | 7957 | 13430 | -0.149 | -0.3456 | No | ||
| 80 | CSNK2B | 23008 | 13539 | -0.159 | -0.3508 | No | ||
| 81 | CLMN | 20988 2119 2104 | 13607 | -0.166 | -0.3537 | No | ||
| 82 | ACIN1 | 3319 2807 21832 | 13776 | -0.182 | -0.3620 | No | ||
| 83 | ZNF23 | 6149 | 13936 | -0.199 | -0.3698 | No | ||
| 84 | PER2 | 3940 13881 | 14228 | -0.237 | -0.3845 | No | ||
| 85 | BTBD1 | 13519 | 14303 | -0.247 | -0.3875 | No | ||
| 86 | HTATIP | 3690 | 14713 | -0.337 | -0.4082 | No | ||
| 87 | C12ORF10 | 2223 2291 | 14933 | -0.395 | -0.4183 | No | ||
| 88 | RPS6KA3 | 8490 | 14980 | -0.408 | -0.4191 | No | ||
| 89 | ZDHHC5 | 14547 | 15079 | -0.445 | -0.4225 | No | ||
| 90 | SART3 | 16428 | 15097 | -0.453 | -0.4215 | No | ||
| 91 | RPL32 | 9743 | 15147 | -0.472 | -0.4221 | No | ||
| 92 | EXOSC3 | 15890 | 15354 | -0.564 | -0.4308 | No | ||
| 93 | B3GAT3 | 23938 | 15383 | -0.578 | -0.4299 | No | ||
| 94 | CLDN12 | 16925 | 15445 | -0.626 | -0.4305 | No | ||
| 95 | SF3B4 | 22269 | 15726 | -0.807 | -0.4422 | No | ||
| 96 | PPP1R11 | 8025 | 15815 | -0.870 | -0.4433 | No | ||
| 97 | ACTB | 8534 337 337 338 | 15929 | -0.962 | -0.4453 | No | ||
| 98 | CHERP | 11367 | 16081 | -1.040 | -0.4490 | Yes | ||
| 99 | PCBP1 | 17085 | 16099 | -1.053 | -0.4454 | Yes | ||
| 100 | VPS16 | 2927 14845 2825 | 16105 | -1.061 | -0.4412 | Yes | ||
| 101 | DDOST | 16021 | 16255 | -1.186 | -0.4442 | Yes | ||
| 102 | NKIRAS2 | 12979 | 16291 | -1.219 | -0.4409 | Yes | ||
| 103 | NRAS | 5191 | 16449 | -1.395 | -0.4434 | Yes | ||
| 104 | CDK9 | 14619 | 16469 | -1.426 | -0.4383 | Yes | ||
| 105 | NUP155 | 2298 5027 | 16563 | -1.521 | -0.4369 | Yes | ||
| 106 | SF3A1 | 7450 | 16568 | -1.527 | -0.4306 | Yes | ||
| 107 | AP1M1 | 18571 | 16582 | -1.540 | -0.4247 | Yes | ||
| 108 | DTX2 | 16674 3488 | 16595 | -1.548 | -0.4187 | Yes | ||
| 109 | PHF6 | 2648 24340 | 16655 | -1.589 | -0.4151 | Yes | ||
| 110 | TBP | 671 1554 | 16689 | -1.625 | -0.4100 | Yes | ||
| 111 | MRPS21 | 12323 | 16701 | -1.635 | -0.4036 | Yes | ||
| 112 | DDX1 | 21104 | 16734 | -1.668 | -0.3982 | Yes | ||
| 113 | MTPN | 17183 | 16779 | -1.716 | -0.3933 | Yes | ||
| 114 | SNIP1 | 16085 | 16847 | -1.807 | -0.3892 | Yes | ||
| 115 | COX6A1 | 8774 4553 | 16863 | -1.830 | -0.3822 | Yes | ||
| 116 | ZNF35 | 19258 | 16903 | -1.887 | -0.3762 | Yes | ||
| 117 | TAF5L | 18712 | 16935 | -1.937 | -0.3696 | Yes | ||
| 118 | MYOHD1 | 20724 | 16952 | -1.961 | -0.3621 | Yes | ||
| 119 | TAF10 | 10785 | 16990 | -2.022 | -0.3555 | Yes | ||
| 120 | PSMB1 | 23118 | 17038 | -2.079 | -0.3491 | Yes | ||
| 121 | CSTF1 | 12515 | 17048 | -2.098 | -0.3407 | Yes | ||
| 122 | RHOT2 | 1533 10073 | 17102 | -2.177 | -0.3342 | Yes | ||
| 123 | PCBP4 | 12235 | 17220 | -2.333 | -0.3306 | Yes | ||
| 124 | EIF3S3 | 12652 | 17269 | -2.460 | -0.3227 | Yes | ||
| 125 | ZNF24 | 7207 | 17307 | -2.540 | -0.3139 | Yes | ||
| 126 | IDH3G | 24132 | 17368 | -2.647 | -0.3058 | Yes | ||
| 127 | BANF1 | 6216 10706 3760 | 17446 | -2.797 | -0.2980 | Yes | ||
| 128 | HTF9C | 4886 1678 | 17479 | -2.864 | -0.2875 | Yes | ||
| 129 | TOMM40 | 6997 | 17562 | -3.010 | -0.2791 | Yes | ||
| 130 | PIGF | 22879 | 17568 | -3.022 | -0.2665 | Yes | ||
| 131 | HSPC171 | 18494 | 17574 | -3.032 | -0.2538 | Yes | ||
| 132 | SPG21 | 19401 | 17576 | -3.035 | -0.2409 | Yes | ||
| 133 | EBNA1BP2 | 8112 7599 12770 | 17665 | -3.236 | -0.2318 | Yes | ||
| 134 | SUV39H1 | 24194 | 17743 | -3.415 | -0.2214 | Yes | ||
| 135 | TOMM22 | 5863 | 17744 | -3.415 | -0.2068 | Yes | ||
| 136 | DNAJC7 | 20227 | 17784 | -3.518 | -0.1939 | Yes | ||
| 137 | SUPT16H | 8541 | 17882 | -3.805 | -0.1829 | Yes | ||
| 138 | SF3B3 | 18746 | 17915 | -3.934 | -0.1678 | Yes | ||
| 139 | BAG4 | 7431 | 17942 | -4.019 | -0.1520 | Yes | ||
| 140 | SLC30A7 | 15186 | 18085 | -4.530 | -0.1404 | Yes | ||
| 141 | PRKAG2 | 24 | 18124 | -4.685 | -0.1224 | Yes | ||
| 142 | SLC39A6 | 23480 | 18314 | -5.451 | -0.1094 | Yes | ||
| 143 | LZIC | 15988 | 18315 | -5.457 | -0.0861 | Yes | ||
| 144 | EIF5A | 11345 20379 6590 | 18392 | -5.951 | -0.0648 | Yes | ||
| 145 | EXTL2 | 15440 | 18514 | -7.089 | -0.0410 | Yes | ||
| 146 | MRPL3 | 3116 19335 | 18611 | -10.888 | 0.0003 | Yes |