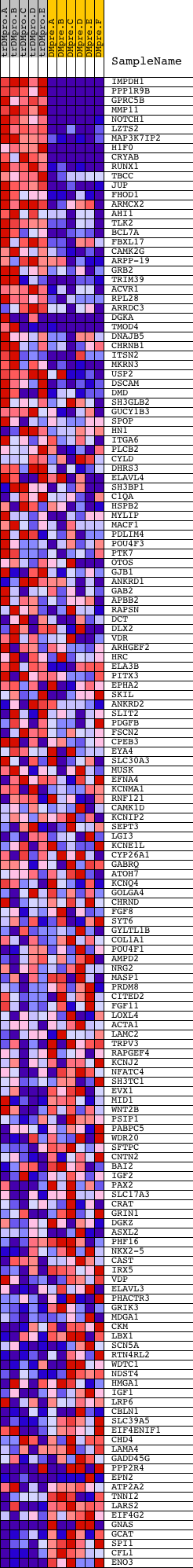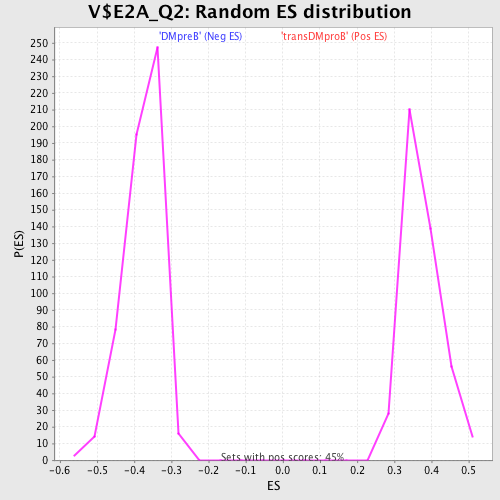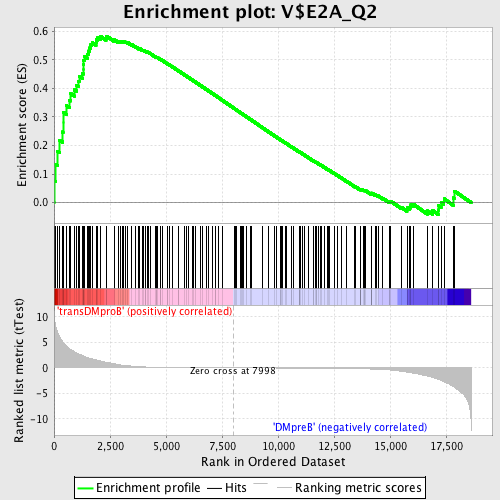
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$E2A_Q2 |
| Enrichment Score (ES) | 0.58276725 |
| Normalized Enrichment Score (NES) | 1.5618892 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.05964831 |
| FWER p-Value | 0.481 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | IMPDH1 | 17197 1131 | 3 | 10.857 | 0.0769 | Yes | ||
| 2 | PPP1R9B | 20698 | 54 | 8.508 | 0.1345 | Yes | ||
| 3 | GPRC5B | 17662 | 148 | 7.065 | 0.1796 | Yes | ||
| 4 | MMP11 | 19727 | 234 | 6.219 | 0.2192 | Yes | ||
| 5 | NOTCH1 | 14649 | 381 | 5.091 | 0.2474 | Yes | ||
| 6 | LZTS2 | 10362 | 401 | 4.974 | 0.2817 | Yes | ||
| 7 | MAP3K7IP2 | 19827 | 402 | 4.966 | 0.3169 | Yes | ||
| 8 | H1F0 | 22428 | 551 | 4.314 | 0.3395 | Yes | ||
| 9 | CRYAB | 19457 | 694 | 3.726 | 0.3582 | Yes | ||
| 10 | RUNX1 | 4481 | 743 | 3.588 | 0.3811 | Yes | ||
| 11 | TBCC | 23215 | 897 | 3.189 | 0.3954 | Yes | ||
| 12 | JUP | 4937 | 1004 | 2.930 | 0.4105 | Yes | ||
| 13 | FHOD1 | 18772 | 1084 | 2.741 | 0.4257 | Yes | ||
| 14 | ARMCX2 | 12530 | 1120 | 2.678 | 0.4428 | Yes | ||
| 15 | AHI1 | 20077 | 1271 | 2.373 | 0.4515 | Yes | ||
| 16 | TLK2 | 20628 | 1299 | 2.326 | 0.4665 | Yes | ||
| 17 | BCL7A | 8063 | 1302 | 2.324 | 0.4829 | Yes | ||
| 18 | FBXL17 | 22910 | 1311 | 2.304 | 0.4988 | Yes | ||
| 19 | CAMK2G | 21905 | 1352 | 2.227 | 0.5124 | Yes | ||
| 20 | ARPP-19 | 19379 | 1494 | 2.002 | 0.5190 | Yes | ||
| 21 | GRB2 | 20149 | 1534 | 1.943 | 0.5307 | Yes | ||
| 22 | TRIM39 | 22994 | 1585 | 1.878 | 0.5413 | Yes | ||
| 23 | ACVR1 | 4334 | 1627 | 1.818 | 0.5520 | Yes | ||
| 24 | RPL28 | 5392 | 1691 | 1.736 | 0.5609 | Yes | ||
| 25 | ARRDC3 | 4191 | 1871 | 1.545 | 0.5622 | Yes | ||
| 26 | DGKA | 3359 19589 | 1891 | 1.523 | 0.5719 | Yes | ||
| 27 | TMOD4 | 1816 1884 15509 | 1950 | 1.461 | 0.5792 | Yes | ||
| 28 | DNAJB5 | 16236 | 2083 | 1.327 | 0.5814 | Yes | ||
| 29 | CHRNB1 | 20384 | 2318 | 1.100 | 0.5766 | Yes | ||
| 30 | ITSN2 | 21327 | 2346 | 1.082 | 0.5828 | Yes | ||
| 31 | MKRN3 | 17810 | 2675 | 0.807 | 0.5707 | No | ||
| 32 | USP2 | 19480 3052 3043 | 2862 | 0.641 | 0.5652 | No | ||
| 33 | DSCAM | 1667 4641 22530 | 2958 | 0.578 | 0.5642 | No | ||
| 34 | DMD | 24295 2647 | 3054 | 0.526 | 0.5627 | No | ||
| 35 | SH3GLB2 | 14628 | 3088 | 0.507 | 0.5645 | No | ||
| 36 | GUCY1B3 | 15311 | 3192 | 0.453 | 0.5622 | No | ||
| 37 | SPOP | 5498 | 3270 | 0.417 | 0.5610 | No | ||
| 38 | HN1 | 9101 | 3459 | 0.336 | 0.5532 | No | ||
| 39 | ITGA6 | 4930 | 3634 | 0.274 | 0.5457 | No | ||
| 40 | PLCB2 | 5262 | 3787 | 0.233 | 0.5391 | No | ||
| 41 | CYLD | 18532 | 3798 | 0.230 | 0.5402 | No | ||
| 42 | DHRS3 | 15997 | 3946 | 0.193 | 0.5336 | No | ||
| 43 | ELAVL4 | 15805 4889 9137 | 3984 | 0.185 | 0.5329 | No | ||
| 44 | SH3BP1 | 632 22431 | 4075 | 0.167 | 0.5292 | No | ||
| 45 | C1QA | 8666 | 4087 | 0.166 | 0.5298 | No | ||
| 46 | HSPB2 | 19121 | 4150 | 0.155 | 0.5275 | No | ||
| 47 | MYLIP | 21652 3189 | 4218 | 0.145 | 0.5249 | No | ||
| 48 | MACF1 | 4316 15763 2382 15764 | 4282 | 0.137 | 0.5225 | No | ||
| 49 | PDLIM4 | 20455 | 4514 | 0.106 | 0.5107 | No | ||
| 50 | POU4F3 | 9604 | 4562 | 0.099 | 0.5089 | No | ||
| 51 | PTK7 | 22959 | 4569 | 0.098 | 0.5092 | No | ||
| 52 | OTOS | 13877 | 4607 | 0.095 | 0.5079 | No | ||
| 53 | GJB1 | 24276 | 4744 | 0.083 | 0.5011 | No | ||
| 54 | ANKRD1 | 23689 | 4758 | 0.082 | 0.5010 | No | ||
| 55 | GAB2 | 1821 18184 2025 | 4834 | 0.077 | 0.4975 | No | ||
| 56 | APBB2 | 8607 | 5045 | 0.066 | 0.4866 | No | ||
| 57 | RAPSN | 2879 14951 | 5075 | 0.064 | 0.4855 | No | ||
| 58 | DCT | 4603 21723 | 5131 | 0.061 | 0.4829 | No | ||
| 59 | DLX2 | 14561 353 | 5282 | 0.055 | 0.4752 | No | ||
| 60 | VDR | 5852 10285 | 5545 | 0.045 | 0.4613 | No | ||
| 61 | ARHGEF2 | 4987 | 5563 | 0.044 | 0.4607 | No | ||
| 62 | HRC | 18251 | 5839 | 0.037 | 0.4461 | No | ||
| 63 | ELA3B | 12592 7494 | 5910 | 0.035 | 0.4425 | No | ||
| 64 | PITX3 | 9571 | 6005 | 0.033 | 0.4377 | No | ||
| 65 | EPHA2 | 16006 | 6188 | 0.029 | 0.4280 | No | ||
| 66 | SKIL | 15617 | 6224 | 0.028 | 0.4263 | No | ||
| 67 | ANKRD2 | 23853 | 6320 | 0.026 | 0.4214 | No | ||
| 68 | SLIT2 | 5456 | 6532 | 0.022 | 0.4101 | No | ||
| 69 | PDGFB | 22199 2311 | 6625 | 0.020 | 0.4052 | No | ||
| 70 | FSCN2 | 20569 | 6809 | 0.017 | 0.3954 | No | ||
| 71 | CPEB3 | 5539 | 6897 | 0.015 | 0.3908 | No | ||
| 72 | EYA4 | 8937 | 7050 | 0.013 | 0.3827 | No | ||
| 73 | SLC30A3 | 5995 | 7183 | 0.011 | 0.3756 | No | ||
| 74 | MUSK | 5199 | 7219 | 0.011 | 0.3738 | No | ||
| 75 | EFNA4 | 1866 15277 | 7324 | 0.009 | 0.3682 | No | ||
| 76 | KCNMA1 | 4947 | 7527 | 0.006 | 0.3573 | No | ||
| 77 | RNF121 | 7952 373 | 8036 | -0.000 | 0.3298 | No | ||
| 78 | CAMK1D | 5977 2948 | 8118 | -0.002 | 0.3254 | No | ||
| 79 | KCNIP2 | 23655 3725 | 8158 | -0.002 | 0.3233 | No | ||
| 80 | SEPT3 | 6275 10775 | 8303 | -0.004 | 0.3156 | No | ||
| 81 | LGI3 | 21967 | 8372 | -0.005 | 0.3119 | No | ||
| 82 | KCNE1L | 24044 | 8427 | -0.006 | 0.3090 | No | ||
| 83 | CYP26A1 | 23871 3695 | 8466 | -0.006 | 0.3070 | No | ||
| 84 | GABRQ | 24315 | 8574 | -0.008 | 0.3013 | No | ||
| 85 | ATOH7 | 20000 | 8765 | -0.010 | 0.2911 | No | ||
| 86 | KCNQ4 | 12247 2533 | 8829 | -0.011 | 0.2877 | No | ||
| 87 | GOLGA4 | 12022 | 9317 | -0.018 | 0.2615 | No | ||
| 88 | CHRND | 3931 4084 | 9589 | -0.022 | 0.2470 | No | ||
| 89 | FGF8 | 23656 | 9837 | -0.026 | 0.2338 | No | ||
| 90 | SYT6 | 13437 7042 12040 | 9929 | -0.027 | 0.2290 | No | ||
| 91 | GYLTL1B | 2969 14520 | 10098 | -0.030 | 0.2201 | No | ||
| 92 | COL1A1 | 8771 | 10126 | -0.031 | 0.2189 | No | ||
| 93 | POU4F1 | 21727 | 10140 | -0.031 | 0.2184 | No | ||
| 94 | AMPD2 | 1931 15197 | 10189 | -0.032 | 0.2160 | No | ||
| 95 | NRG2 | 4274 | 10326 | -0.034 | 0.2089 | No | ||
| 96 | MASP1 | 1632 22626 | 10375 | -0.035 | 0.2065 | No | ||
| 97 | PRDM8 | 8084 | 10613 | -0.039 | 0.1940 | No | ||
| 98 | CITED2 | 5118 14477 | 10616 | -0.039 | 0.1942 | No | ||
| 99 | FGF11 | 20383 | 10697 | -0.040 | 0.1901 | No | ||
| 100 | LOXL4 | 23672 | 10932 | -0.045 | 0.1777 | No | ||
| 101 | ACTA1 | 18715 | 10949 | -0.045 | 0.1772 | No | ||
| 102 | LAMC2 | 4983 9266 13806 3975 | 10971 | -0.046 | 0.1764 | No | ||
| 103 | TRPV3 | 20788 | 11020 | -0.047 | 0.1741 | No | ||
| 104 | RAPGEF4 | 12141 | 11078 | -0.048 | 0.1714 | No | ||
| 105 | KCNJ2 | 20612 | 11193 | -0.050 | 0.1655 | No | ||
| 106 | NFATC4 | 22002 | 11337 | -0.054 | 0.1582 | No | ||
| 107 | SH3TC1 | 16554 | 11563 | -0.059 | 0.1464 | No | ||
| 108 | EVX1 | 17442 | 11586 | -0.059 | 0.1456 | No | ||
| 109 | MID1 | 5097 5098 | 11671 | -0.062 | 0.1415 | No | ||
| 110 | WNT2B | 15214 1838 | 11702 | -0.063 | 0.1404 | No | ||
| 111 | PSIP1 | 4159 2507 8319 | 11710 | -0.063 | 0.1404 | No | ||
| 112 | PABPC5 | 24077 | 11815 | -0.066 | 0.1353 | No | ||
| 113 | WDR20 | 21258 | 11870 | -0.068 | 0.1328 | No | ||
| 114 | SFTPC | 21759 | 11919 | -0.070 | 0.1307 | No | ||
| 115 | CNTN2 | 5627 | 12091 | -0.075 | 0.1220 | No | ||
| 116 | BAI2 | 2541 2379 16067 | 12224 | -0.080 | 0.1154 | No | ||
| 117 | IGF2 | 17550 | 12252 | -0.082 | 0.1145 | No | ||
| 118 | PAX2 | 9530 24416 | 12304 | -0.085 | 0.1123 | No | ||
| 119 | SLC17A3 | 3165 21694 | 12509 | -0.093 | 0.1020 | No | ||
| 120 | CRAT | 4557 2804 2896 | 12668 | -0.100 | 0.0941 | No | ||
| 121 | GRIN1 | 9041 4804 | 12815 | -0.108 | 0.0870 | No | ||
| 122 | DGKZ | 2836 14522 | 13068 | -0.123 | 0.0742 | No | ||
| 123 | ASXL2 | 7957 | 13430 | -0.149 | 0.0557 | No | ||
| 124 | PHF16 | 11795 6901 | 13461 | -0.151 | 0.0551 | No | ||
| 125 | NKX2-5 | 23060 | 13668 | -0.171 | 0.0452 | No | ||
| 126 | CAST | 3182 4478 3177 | 13673 | -0.172 | 0.0462 | No | ||
| 127 | IRX5 | 18527 | 13674 | -0.172 | 0.0474 | No | ||
| 128 | VDP | 16794 | 13697 | -0.174 | 0.0475 | No | ||
| 129 | ELAVL3 | 9136 | 13814 | -0.186 | 0.0425 | No | ||
| 130 | PHACTR3 | 7900 2770 | 13843 | -0.190 | 0.0423 | No | ||
| 131 | GRIK3 | 16083 | 13875 | -0.193 | 0.0420 | No | ||
| 132 | MDGA1 | 13231 | 13898 | -0.194 | 0.0422 | No | ||
| 133 | CKM | 18362 | 14156 | -0.227 | 0.0299 | No | ||
| 134 | LBX1 | 98 23659 | 14171 | -0.229 | 0.0308 | No | ||
| 135 | SCN5A | 5412 | 14176 | -0.229 | 0.0322 | No | ||
| 136 | RTN4RL2 | 14544 | 14178 | -0.230 | 0.0337 | No | ||
| 137 | WDTC1 | 10512 | 14331 | -0.252 | 0.0273 | No | ||
| 138 | NDST4 | 12259 7228 | 14389 | -0.262 | 0.0261 | No | ||
| 139 | HMGA1 | 23323 | 14469 | -0.276 | 0.0237 | No | ||
| 140 | IGF1 | 3352 9156 3409 | 14678 | -0.326 | 0.0148 | No | ||
| 141 | LRP6 | 9286 | 14965 | -0.404 | 0.0022 | No | ||
| 142 | CBLN1 | 18800 | 14989 | -0.411 | 0.0038 | No | ||
| 143 | SLC39A5 | 3364 19596 | 15019 | -0.420 | 0.0052 | No | ||
| 144 | EIF4ENIF1 | 20971 | 15488 | -0.648 | -0.0155 | No | ||
| 145 | CHD4 | 8418 17281 4225 | 15754 | -0.826 | -0.0240 | No | ||
| 146 | LAMA4 | 20054 | 15757 | -0.827 | -0.0182 | No | ||
| 147 | GADD45G | 21637 | 15858 | -0.901 | -0.0173 | No | ||
| 148 | PPP2R4 | 15050 2910 | 15891 | -0.926 | -0.0124 | No | ||
| 149 | EPN2 | 20413 945 946 955 | 15928 | -0.962 | -0.0075 | No | ||
| 150 | ATP2A2 | 4421 3481 | 16060 | -1.027 | -0.0074 | No | ||
| 151 | TNNI2 | 18005 | 16666 | -1.600 | -0.0288 | No | ||
| 152 | LARS2 | 19255 | 16889 | -1.870 | -0.0275 | No | ||
| 153 | EIF4G2 | 1908 8892 | 17147 | -2.222 | -0.0257 | No | ||
| 154 | GNAS | 9025 2963 2752 | 17156 | -2.230 | -0.0103 | No | ||
| 155 | GCAT | 11234 | 17278 | -2.477 | 0.0007 | No | ||
| 156 | SPI1 | 14949 | 17404 | -2.714 | 0.0132 | No | ||
| 157 | CFL1 | 4516 | 17815 | -3.604 | 0.0166 | No | ||
| 158 | ENO3 | 8905 | 17874 | -3.773 | 0.0402 | No |