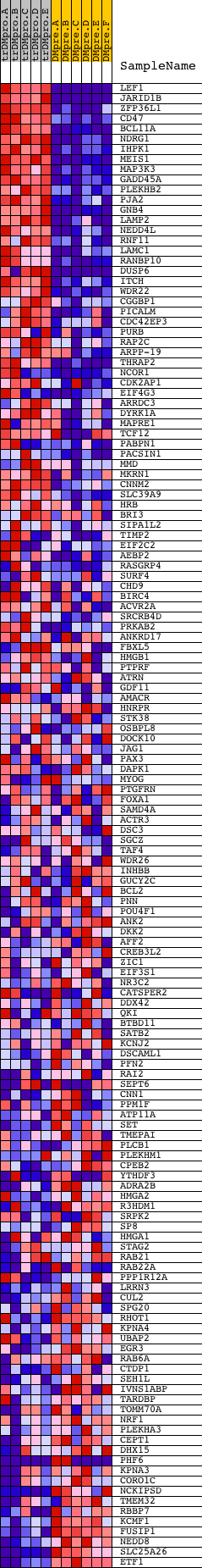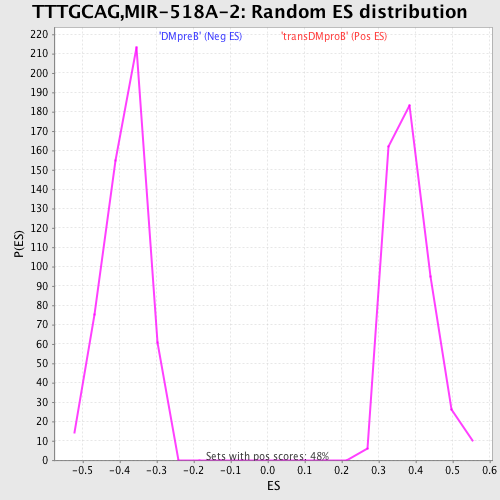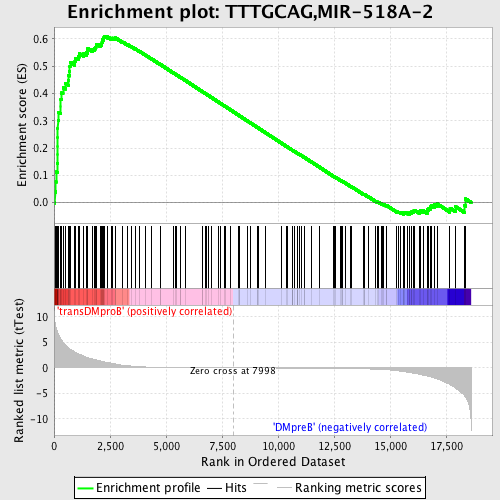
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | TTTGCAG,MIR-518A-2 |
| Enrichment Score (ES) | 0.61032766 |
| Normalized Enrichment Score (NES) | 1.596586 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.05206013 |
| FWER p-Value | 0.334 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | LEF1 | 1860 15420 | 39 | 8.951 | 0.0398 | Yes | ||
| 2 | JARID1B | 14123 | 75 | 8.103 | 0.0758 | Yes | ||
| 3 | ZFP36L1 | 4453 4454 | 88 | 7.945 | 0.1124 | Yes | ||
| 4 | CD47 | 4933 | 143 | 7.118 | 0.1428 | Yes | ||
| 5 | BCL11A | 4691 | 159 | 6.951 | 0.1745 | Yes | ||
| 6 | NDRG1 | 22276 | 161 | 6.937 | 0.2070 | Yes | ||
| 7 | IHPK1 | 11336 | 163 | 6.911 | 0.2393 | Yes | ||
| 8 | MEIS1 | 20524 | 167 | 6.817 | 0.2710 | Yes | ||
| 9 | MAP3K3 | 20626 | 173 | 6.715 | 0.3022 | Yes | ||
| 10 | GADD45A | 17129 | 198 | 6.482 | 0.3313 | Yes | ||
| 11 | PLEKHB2 | 10390 | 300 | 5.626 | 0.3521 | Yes | ||
| 12 | PJA2 | 5904 | 301 | 5.616 | 0.3784 | Yes | ||
| 13 | GNB4 | 4787 9028 | 334 | 5.403 | 0.4020 | Yes | ||
| 14 | LAMP2 | 9267 2653 | 398 | 4.984 | 0.4219 | Yes | ||
| 15 | NEDD4L | 8192 | 510 | 4.462 | 0.4368 | Yes | ||
| 16 | RNF11 | 11383 2509 | 636 | 3.942 | 0.4485 | Yes | ||
| 17 | LAMC1 | 13805 | 653 | 3.867 | 0.4658 | Yes | ||
| 18 | RANBP10 | 18764 | 686 | 3.745 | 0.4816 | Yes | ||
| 19 | DUSP6 | 19891 3399 | 703 | 3.694 | 0.4980 | Yes | ||
| 20 | ITCH | 4928 9185 | 718 | 3.648 | 0.5143 | Yes | ||
| 21 | WDR22 | 6803 | 927 | 3.113 | 0.5176 | Yes | ||
| 22 | CGGBP1 | 22724 | 963 | 3.008 | 0.5298 | Yes | ||
| 23 | PICALM | 18191 | 1089 | 2.731 | 0.5358 | Yes | ||
| 24 | CDC42EP3 | 6436 | 1130 | 2.659 | 0.5461 | Yes | ||
| 25 | PURB | 5335 | 1325 | 2.278 | 0.5463 | Yes | ||
| 26 | RAP2C | 24156 | 1439 | 2.082 | 0.5499 | Yes | ||
| 27 | ARPP-19 | 19379 | 1494 | 2.002 | 0.5564 | Yes | ||
| 28 | THRAP2 | 8009 | 1504 | 1.991 | 0.5652 | Yes | ||
| 29 | NCOR1 | 5402 | 1723 | 1.699 | 0.5614 | Yes | ||
| 30 | CDK2AP1 | 8858 | 1792 | 1.633 | 0.5653 | Yes | ||
| 31 | EIF4G3 | 10517 | 1837 | 1.584 | 0.5704 | Yes | ||
| 32 | ARRDC3 | 4191 | 1871 | 1.545 | 0.5758 | Yes | ||
| 33 | DYRK1A | 4649 | 1900 | 1.517 | 0.5814 | Yes | ||
| 34 | MAPRE1 | 4652 | 2057 | 1.347 | 0.5793 | Yes | ||
| 35 | TCF12 | 10044 3125 | 2134 | 1.282 | 0.5812 | Yes | ||
| 36 | PABPN1 | 12018 | 2135 | 1.279 | 0.5871 | Yes | ||
| 37 | PACSIN1 | 6257 23322 10744 | 2152 | 1.266 | 0.5922 | Yes | ||
| 38 | MMD | 20706 | 2158 | 1.260 | 0.5978 | Yes | ||
| 39 | MKRN1 | 17178 | 2202 | 1.215 | 0.6012 | Yes | ||
| 40 | CNNM2 | 8251 | 2222 | 1.194 | 0.6058 | Yes | ||
| 41 | SLC39A9 | 11597 | 2240 | 1.172 | 0.6103 | Yes | ||
| 42 | HRB | 14208 4066 | 2374 | 1.056 | 0.6081 | No | ||
| 43 | BRI3 | 15101 | 2565 | 0.904 | 0.6020 | No | ||
| 44 | SIPA1L2 | 10863 | 2614 | 0.858 | 0.6034 | No | ||
| 45 | TIMP2 | 10178 | 2718 | 0.769 | 0.6015 | No | ||
| 46 | EIF2C2 | 6247 | 2735 | 0.755 | 0.6041 | No | ||
| 47 | AEBP2 | 4359 | 3063 | 0.518 | 0.5889 | No | ||
| 48 | RASGRP4 | 6119 | 3284 | 0.411 | 0.5789 | No | ||
| 49 | SURF4 | 14646 | 3432 | 0.348 | 0.5725 | No | ||
| 50 | CHD9 | 6784 11553 18531 | 3617 | 0.279 | 0.5639 | No | ||
| 51 | BIRC4 | 2638 8609 | 3793 | 0.231 | 0.5555 | No | ||
| 52 | ACVR2A | 8542 | 4076 | 0.167 | 0.5410 | No | ||
| 53 | SRCRB4D | 8459 | 4358 | 0.125 | 0.5264 | No | ||
| 54 | PRKAB2 | 15488 | 4728 | 0.084 | 0.5068 | No | ||
| 55 | ANKRD17 | 13496 3474 8170 | 4736 | 0.084 | 0.5068 | No | ||
| 56 | FBXL5 | 6330 10828 16540 | 5315 | 0.053 | 0.4758 | No | ||
| 57 | HMGB1 | 9094 4855 | 5397 | 0.050 | 0.4716 | No | ||
| 58 | PTPRF | 5331 2545 | 5442 | 0.048 | 0.4695 | No | ||
| 59 | ATRN | 4432 2964 | 5624 | 0.042 | 0.4599 | No | ||
| 60 | GDF11 | 9011 | 5650 | 0.042 | 0.4587 | No | ||
| 61 | AMACR | 22506 | 5854 | 0.036 | 0.4479 | No | ||
| 62 | HNRPR | 13196 7909 2500 | 6617 | 0.020 | 0.4067 | No | ||
| 63 | STK38 | 23045 | 6636 | 0.020 | 0.4058 | No | ||
| 64 | OSBPL8 | 6198 | 6747 | 0.018 | 0.4000 | No | ||
| 65 | DOCK10 | 4004 13904 9970 | 6807 | 0.017 | 0.3968 | No | ||
| 66 | JAG1 | 14415 | 6887 | 0.015 | 0.3926 | No | ||
| 67 | PAX3 | 13910 | 7013 | 0.014 | 0.3859 | No | ||
| 68 | DAPK1 | 7632 | 7345 | 0.009 | 0.3681 | No | ||
| 69 | MYOG | 14129 | 7437 | 0.007 | 0.3632 | No | ||
| 70 | PTGFRN | 15224 | 7620 | 0.005 | 0.3533 | No | ||
| 71 | FOXA1 | 21060 | 7666 | 0.004 | 0.3509 | No | ||
| 72 | SAMD4A | 13211 | 7861 | 0.002 | 0.3404 | No | ||
| 73 | ACTR3 | 13166 | 8219 | -0.003 | 0.3211 | No | ||
| 74 | DSC3 | 23487 | 8254 | -0.004 | 0.3193 | No | ||
| 75 | SGCZ | 6354 | 8290 | -0.004 | 0.3174 | No | ||
| 76 | TAF4 | 14319 | 8626 | -0.008 | 0.2993 | No | ||
| 77 | WDR26 | 13733 | 8751 | -0.010 | 0.2926 | No | ||
| 78 | INHBB | 13858 | 9069 | -0.015 | 0.2756 | No | ||
| 79 | GUCY2C | 1140 16954 | 9096 | -0.015 | 0.2742 | No | ||
| 80 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 9115 | -0.015 | 0.2733 | No | ||
| 81 | PNN | 9596 | 9439 | -0.020 | 0.2559 | No | ||
| 82 | POU4F1 | 21727 | 10140 | -0.031 | 0.2182 | No | ||
| 83 | ANK2 | 4275 | 10354 | -0.035 | 0.2068 | No | ||
| 84 | DKK2 | 15418 | 10429 | -0.036 | 0.2030 | No | ||
| 85 | AFF2 | 8977 | 10626 | -0.039 | 0.1925 | No | ||
| 86 | CREB3L2 | 17182 | 10648 | -0.039 | 0.1916 | No | ||
| 87 | ZIC1 | 19040 | 10649 | -0.039 | 0.1918 | No | ||
| 88 | EIF3S1 | 905 8114 | 10712 | -0.041 | 0.1886 | No | ||
| 89 | NR3C2 | 18562 13798 | 10851 | -0.043 | 0.1813 | No | ||
| 90 | CATSPER2 | 10014 | 10863 | -0.044 | 0.1809 | No | ||
| 91 | DDX42 | 20625 | 10869 | -0.044 | 0.1809 | No | ||
| 92 | QKI | 23128 5340 | 10973 | -0.046 | 0.1755 | No | ||
| 93 | BTBD11 | 13147 | 11042 | -0.047 | 0.1720 | No | ||
| 94 | SATB2 | 5604 | 11162 | -0.050 | 0.1658 | No | ||
| 95 | KCNJ2 | 20612 | 11193 | -0.050 | 0.1645 | No | ||
| 96 | DSCAML1 | 4338 13224 | 11510 | -0.057 | 0.1476 | No | ||
| 97 | PFN2 | 15337 1818 | 11826 | -0.066 | 0.1309 | No | ||
| 98 | RAI2 | 10762 | 12488 | -0.093 | 0.0955 | No | ||
| 99 | SEPT6 | 2591 7132 | 12523 | -0.094 | 0.0941 | No | ||
| 100 | CNN1 | 8759 | 12559 | -0.096 | 0.0927 | No | ||
| 101 | PPM1F | 22841 1716 | 12769 | -0.106 | 0.0818 | No | ||
| 102 | ATP11A | 18683 6934 | 12837 | -0.110 | 0.0787 | No | ||
| 103 | SET | 7070 | 12853 | -0.110 | 0.0784 | No | ||
| 104 | TMEPAI | 51 | 12889 | -0.112 | 0.0771 | No | ||
| 105 | PLCB1 | 14832 2821 | 13019 | -0.120 | 0.0707 | No | ||
| 106 | PLEKHM1 | 20196 1449 | 13246 | -0.135 | 0.0590 | No | ||
| 107 | CPEB2 | 6077 | 13296 | -0.139 | 0.0570 | No | ||
| 108 | YTHDF3 | 15630 10469 6019 | 13802 | -0.185 | 0.0306 | No | ||
| 109 | ADRA2B | 8562 4356 14863 | 13845 | -0.190 | 0.0292 | No | ||
| 110 | HMGA2 | 4857 9098 3341 3444 | 13863 | -0.191 | 0.0292 | No | ||
| 111 | R3HDM1 | 4002 974 | 14017 | -0.208 | 0.0219 | No | ||
| 112 | SRPK2 | 5513 | 14351 | -0.255 | 0.0050 | No | ||
| 113 | SP8 | 21122 | 14424 | -0.268 | 0.0024 | No | ||
| 114 | HMGA1 | 23323 | 14469 | -0.276 | 0.0013 | No | ||
| 115 | STAG2 | 5521 | 14608 | -0.308 | -0.0047 | No | ||
| 116 | RAB21 | 5696 | 14679 | -0.326 | -0.0070 | No | ||
| 117 | RAB22A | 2888 5343 | 14719 | -0.338 | -0.0075 | No | ||
| 118 | PPP1R12A | 3354 19886 | 14818 | -0.361 | -0.0111 | No | ||
| 119 | LRRN3 | 21077 | 14836 | -0.366 | -0.0103 | No | ||
| 120 | CUL2 | 23630 | 15304 | -0.536 | -0.0331 | No | ||
| 121 | SPG20 | 6024 | 15391 | -0.583 | -0.0350 | No | ||
| 122 | RHOT1 | 12227 | 15467 | -0.638 | -0.0361 | No | ||
| 123 | KPNA4 | 15321 | 15595 | -0.706 | -0.0397 | No | ||
| 124 | UBAP2 | 15912 | 15603 | -0.711 | -0.0367 | No | ||
| 125 | EGR3 | 4656 | 15650 | -0.741 | -0.0358 | No | ||
| 126 | RAB6A | 18173 3905 | 15767 | -0.834 | -0.0381 | No | ||
| 127 | CTDP1 | 23399 1969 | 15872 | -0.913 | -0.0395 | No | ||
| 128 | SEH1L | 13001 | 15883 | -0.921 | -0.0357 | No | ||
| 129 | IVNS1ABP | 4384 4050 | 15957 | -0.984 | -0.0351 | No | ||
| 130 | TARDBP | 10520 6066 | 15969 | -0.990 | -0.0310 | No | ||
| 131 | TOMM70A | 6608 | 16056 | -1.025 | -0.0309 | No | ||
| 132 | NRF1 | 17499 | 16089 | -1.043 | -0.0277 | No | ||
| 133 | PLEKHA3 | 14978 | 16319 | -1.245 | -0.0343 | No | ||
| 134 | CEPT1 | 13624 | 16348 | -1.275 | -0.0298 | No | ||
| 135 | DHX15 | 8842 | 16474 | -1.430 | -0.0299 | No | ||
| 136 | PHF6 | 2648 24340 | 16655 | -1.589 | -0.0322 | No | ||
| 137 | KPNA3 | 21797 | 16656 | -1.591 | -0.0248 | No | ||
| 138 | CORO1C | 16425 | 16728 | -1.662 | -0.0208 | No | ||
| 139 | NCKIPSD | 19306 | 16789 | -1.734 | -0.0160 | No | ||
| 140 | TMEM32 | 24149 | 16849 | -1.812 | -0.0107 | No | ||
| 141 | RBBP7 | 10887 6366 10886 | 16974 | -1.995 | -0.0080 | No | ||
| 142 | KCMF1 | 17112 | 17100 | -2.175 | -0.0046 | No | ||
| 143 | FUSIP1 | 4715 16036 | 17671 | -3.254 | -0.0202 | No | ||
| 144 | NEDD8 | 21822 | 17925 | -3.969 | -0.0154 | No | ||
| 145 | SLC25A26 | 12552 | 18321 | -5.502 | -0.0110 | No | ||
| 146 | ETF1 | 23467 | 18367 | -5.743 | 0.0135 | No |