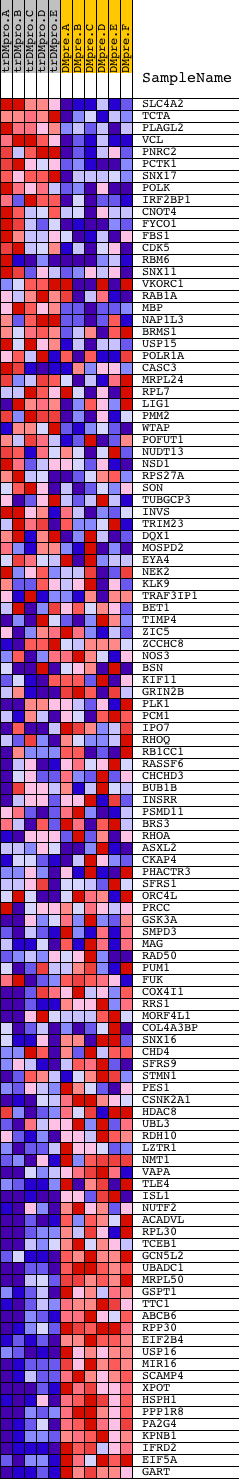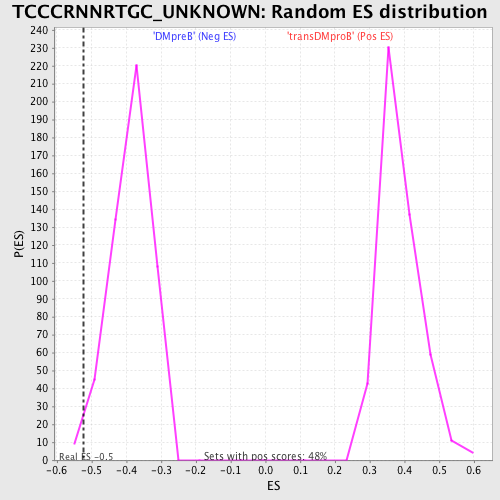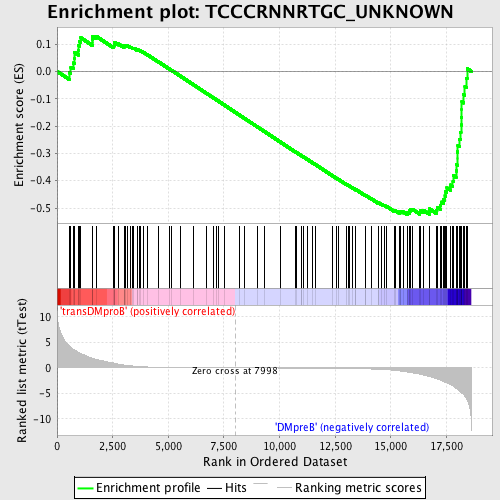
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | TCCCRNNRTGC_UNKNOWN |
| Enrichment Score (ES) | -0.52364916 |
| Normalized Enrichment Score (NES) | -1.3529438 |
| Nominal p-value | 0.015503876 |
| FDR q-value | 0.39106807 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SLC4A2 | 9828 | 552 | 4.310 | -0.0046 | No | ||
| 2 | TCTA | 4167 | 619 | 4.015 | 0.0154 | No | ||
| 3 | PLAGL2 | 7055 | 726 | 3.636 | 0.0310 | No | ||
| 4 | VCL | 22083 | 775 | 3.518 | 0.0490 | No | ||
| 5 | PNRC2 | 15709 | 784 | 3.499 | 0.0691 | No | ||
| 6 | PCTK1 | 24369 | 961 | 3.013 | 0.0773 | No | ||
| 7 | SNX17 | 3495 16885 | 970 | 3.004 | 0.0945 | No | ||
| 8 | POLK | 21384 | 1006 | 2.924 | 0.1097 | No | ||
| 9 | IRF2BP1 | 18368 | 1066 | 2.792 | 0.1229 | No | ||
| 10 | CNOT4 | 7010 1022 | 1581 | 1.884 | 0.1062 | No | ||
| 11 | FYCO1 | 18952 | 1583 | 1.878 | 0.1171 | No | ||
| 12 | FBS1 | 18067 | 1595 | 1.869 | 0.1275 | No | ||
| 13 | CDK5 | 16591 | 1747 | 1.669 | 0.1291 | No | ||
| 14 | RBM6 | 9707 3095 3063 2995 | 2523 | 0.944 | 0.0928 | No | ||
| 15 | SNX11 | 13210 | 2559 | 0.917 | 0.0963 | No | ||
| 16 | VKORC1 | 17607 2126 | 2581 | 0.893 | 0.1004 | No | ||
| 17 | RAB1A | 20942 | 2584 | 0.892 | 0.1055 | No | ||
| 18 | MBP | 23502 | 2751 | 0.738 | 0.1008 | No | ||
| 19 | NAP1L3 | 24266 | 3011 | 0.549 | 0.0901 | No | ||
| 20 | BRMS1 | 8403 | 3013 | 0.549 | 0.0932 | No | ||
| 21 | USP15 | 4758 3377 | 3023 | 0.544 | 0.0959 | No | ||
| 22 | POLR1A | 9749 5393 | 3076 | 0.513 | 0.0961 | No | ||
| 23 | CASC3 | 9643 | 3166 | 0.463 | 0.0940 | No | ||
| 24 | MRPL24 | 15556 | 3302 | 0.399 | 0.0891 | No | ||
| 25 | RPL7 | 9748 | 3408 | 0.356 | 0.0855 | No | ||
| 26 | LIG1 | 18388 1749 1493 | 3446 | 0.342 | 0.0855 | No | ||
| 27 | PMM2 | 1670 1666 1707 22863 1756 | 3609 | 0.282 | 0.0784 | No | ||
| 28 | WTAP | 7215 | 3611 | 0.281 | 0.0800 | No | ||
| 29 | POFUT1 | 4697 | 3687 | 0.259 | 0.0775 | No | ||
| 30 | NUDT13 | 22088 | 3765 | 0.239 | 0.0747 | No | ||
| 31 | NSD1 | 2134 5197 | 3895 | 0.206 | 0.0689 | No | ||
| 32 | RPS27A | 13433 | 4066 | 0.169 | 0.0607 | No | ||
| 33 | SON | 5473 1657 1684 | 4552 | 0.100 | 0.0351 | No | ||
| 34 | TUBGCP3 | 18933 | 5055 | 0.065 | 0.0084 | No | ||
| 35 | INVS | 2397 16211 | 5124 | 0.062 | 0.0050 | No | ||
| 36 | TRIM23 | 8165 3230 | 5549 | 0.045 | -0.0176 | No | ||
| 37 | DQX1 | 17399 | 6111 | 0.030 | -0.0478 | No | ||
| 38 | MOSPD2 | 24009 | 6708 | 0.018 | -0.0799 | No | ||
| 39 | EYA4 | 8937 | 7050 | 0.013 | -0.0982 | No | ||
| 40 | NEK2 | 5160 | 7143 | 0.012 | -0.1031 | No | ||
| 41 | KLK9 | 18267 | 7267 | 0.010 | -0.1097 | No | ||
| 42 | TRAF3IP1 | 14186 | 7511 | 0.006 | -0.1228 | No | ||
| 43 | BET1 | 17226 | 8189 | -0.003 | -0.1594 | No | ||
| 44 | TIMP4 | 17036 | 8433 | -0.006 | -0.1725 | No | ||
| 45 | ZIC5 | 12268 | 9023 | -0.014 | -0.2042 | No | ||
| 46 | ZCCHC8 | 3570 7696 | 9325 | -0.018 | -0.2204 | No | ||
| 47 | NOS3 | 16906 885 | 10060 | -0.030 | -0.2599 | No | ||
| 48 | BSN | 18997 | 10736 | -0.041 | -0.2961 | No | ||
| 49 | KIF11 | 23873 | 10774 | -0.042 | -0.2979 | No | ||
| 50 | GRIN2B | 16957 | 10968 | -0.046 | -0.3080 | No | ||
| 51 | PLK1 | 9590 5266 | 10982 | -0.046 | -0.3085 | No | ||
| 52 | PCM1 | 3780 5229 | 11059 | -0.047 | -0.3123 | No | ||
| 53 | IPO7 | 6130 | 11240 | -0.051 | -0.3217 | No | ||
| 54 | RHOQ | 23142 | 11493 | -0.057 | -0.3350 | No | ||
| 55 | RB1CC1 | 4486 14299 | 11599 | -0.060 | -0.3403 | No | ||
| 56 | RASSF6 | 16484 | 12368 | -0.087 | -0.3813 | No | ||
| 57 | CHCHD3 | 17188 | 12541 | -0.095 | -0.3901 | No | ||
| 58 | BUB1B | 14908 | 12669 | -0.100 | -0.3964 | No | ||
| 59 | INSRR | 15557 | 13007 | -0.120 | -0.4139 | No | ||
| 60 | PSMD11 | 12772 7600 | 13078 | -0.123 | -0.4169 | No | ||
| 61 | BRS3 | 24333 | 13137 | -0.128 | -0.4193 | No | ||
| 62 | RHOA | 8624 4409 4410 | 13287 | -0.138 | -0.4266 | No | ||
| 63 | ASXL2 | 7957 | 13430 | -0.149 | -0.4334 | No | ||
| 64 | CKAP4 | 19663 | 13432 | -0.149 | -0.4325 | No | ||
| 65 | PHACTR3 | 7900 2770 | 13843 | -0.190 | -0.4536 | No | ||
| 66 | SFRS1 | 8492 | 13847 | -0.190 | -0.4526 | No | ||
| 67 | ORC4L | 11172 6460 | 14118 | -0.221 | -0.4659 | No | ||
| 68 | PRCC | 15298 1924 | 14439 | -0.271 | -0.4816 | No | ||
| 69 | GSK3A | 405 | 14455 | -0.274 | -0.4809 | No | ||
| 70 | SMPD3 | 18757 | 14600 | -0.306 | -0.4868 | No | ||
| 71 | MAG | 17880 | 14699 | -0.332 | -0.4902 | No | ||
| 72 | RAD50 | 1198 20460 | 14815 | -0.361 | -0.4943 | No | ||
| 73 | PUM1 | 8160 | 15157 | -0.478 | -0.5099 | No | ||
| 74 | FUK | 18745 | 15191 | -0.489 | -0.5088 | No | ||
| 75 | COX4I1 | 18444 | 15393 | -0.584 | -0.5163 | No | ||
| 76 | RRS1 | 12223 | 15430 | -0.609 | -0.5146 | No | ||
| 77 | MORF4L1 | 5723 | 15432 | -0.613 | -0.5111 | No | ||
| 78 | COL4A3BP | 3221 7522 | 15566 | -0.691 | -0.5142 | No | ||
| 79 | SNX16 | 15374 | 15741 | -0.815 | -0.5189 | Yes | ||
| 80 | CHD4 | 8418 17281 4225 | 15754 | -0.826 | -0.5147 | Yes | ||
| 81 | SFRS9 | 16731 | 15840 | -0.887 | -0.5141 | Yes | ||
| 82 | STMN1 | 9261 | 15852 | -0.895 | -0.5094 | Yes | ||
| 83 | PES1 | 7236 | 15887 | -0.923 | -0.5058 | Yes | ||
| 84 | CSNK2A1 | 14797 | 15954 | -0.982 | -0.5036 | Yes | ||
| 85 | HDAC8 | 24090 | 16307 | -1.236 | -0.5154 | Yes | ||
| 86 | UBL3 | 16284 | 16325 | -1.248 | -0.5090 | Yes | ||
| 87 | RDH10 | 8272 | 16486 | -1.440 | -0.5092 | Yes | ||
| 88 | LZTR1 | 22835 | 16735 | -1.669 | -0.5129 | Yes | ||
| 89 | NMT1 | 20638 | 16738 | -1.674 | -0.5031 | Yes | ||
| 90 | VAPA | 22908 | 17060 | -2.112 | -0.5081 | Yes | ||
| 91 | TLE4 | 23720 3697 | 17076 | -2.143 | -0.4964 | Yes | ||
| 92 | ISL1 | 21338 | 17228 | -2.358 | -0.4907 | Yes | ||
| 93 | NUTF2 | 12632 7529 | 17255 | -2.416 | -0.4779 | Yes | ||
| 94 | ACADVL | 20375 | 17365 | -2.645 | -0.4683 | Yes | ||
| 95 | RPL30 | 9742 | 17434 | -2.770 | -0.4557 | Yes | ||
| 96 | TCEB1 | 13995 | 17465 | -2.829 | -0.4408 | Yes | ||
| 97 | GCN5L2 | 4763 | 17521 | -2.950 | -0.4264 | Yes | ||
| 98 | UBADC1 | 2962 13608 | 17676 | -3.265 | -0.4156 | Yes | ||
| 99 | MRPL50 | 11372 | 17792 | -3.534 | -0.4011 | Yes | ||
| 100 | GSPT1 | 9046 | 17814 | -3.603 | -0.3811 | Yes | ||
| 101 | TTC1 | 7342 | 17935 | -3.997 | -0.3642 | Yes | ||
| 102 | ABCB6 | 13162 | 17948 | -4.040 | -0.3411 | Yes | ||
| 103 | RPP30 | 23877 | 17995 | -4.175 | -0.3191 | Yes | ||
| 104 | EIF2B4 | 16574 | 17996 | -4.187 | -0.2946 | Yes | ||
| 105 | USP16 | 22716 | 18014 | -4.245 | -0.2706 | Yes | ||
| 106 | MIR16 | 17663 1354 | 18084 | -4.529 | -0.2478 | Yes | ||
| 107 | SCAMP4 | 19941 | 18126 | -4.686 | -0.2225 | Yes | ||
| 108 | XPOT | 13090 | 18156 | -4.804 | -0.1959 | Yes | ||
| 109 | HSPH1 | 16282 | 18166 | -4.828 | -0.1681 | Yes | ||
| 110 | PPP1R8 | 15730 | 18190 | -4.937 | -0.1404 | Yes | ||
| 111 | PA2G4 | 5265 | 18195 | -4.947 | -0.1116 | Yes | ||
| 112 | KPNB1 | 20274 | 18273 | -5.185 | -0.0854 | Yes | ||
| 113 | IFRD2 | 19320 | 18331 | -5.544 | -0.0559 | Yes | ||
| 114 | EIF5A | 11345 20379 6590 | 18392 | -5.951 | -0.0243 | Yes | ||
| 115 | GART | 22543 1754 | 18431 | -6.196 | 0.0100 | Yes |