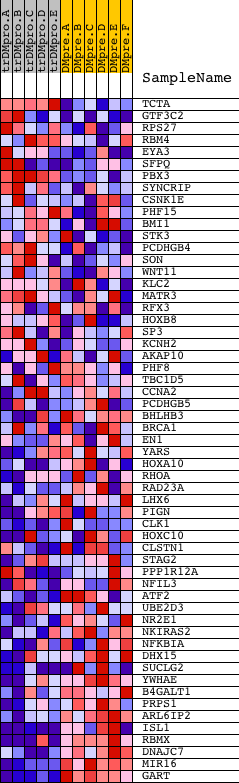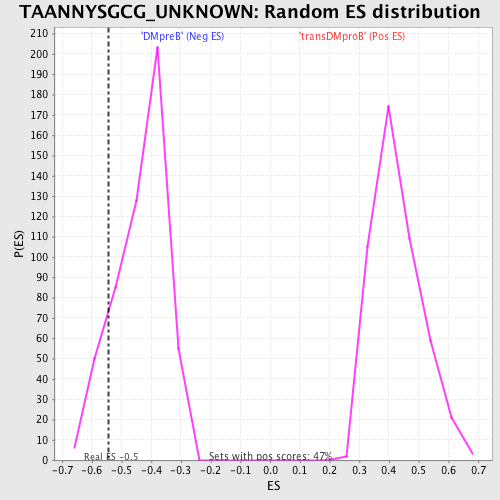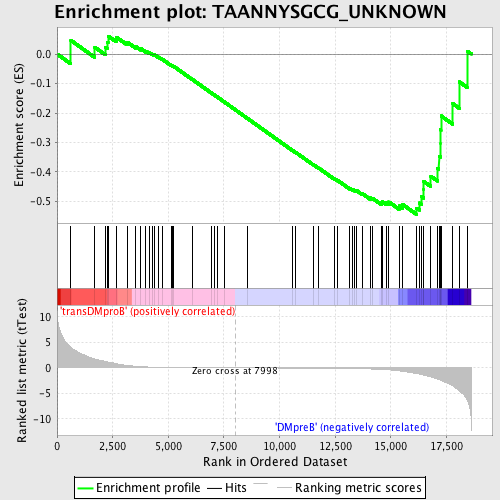
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | TAANNYSGCG_UNKNOWN |
| Enrichment Score (ES) | -0.5441344 |
| Normalized Enrichment Score (NES) | -1.2540388 |
| Nominal p-value | 0.12713473 |
| FDR q-value | 0.6475328 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TCTA | 4167 | 619 | 4.015 | 0.0464 | No | ||
| 2 | GTF3C2 | 7749 | 1695 | 1.732 | 0.0228 | No | ||
| 3 | RPS27 | 12183 | 2173 | 1.245 | 0.0218 | No | ||
| 4 | RBM4 | 9706 3693 | 2253 | 1.162 | 0.0407 | No | ||
| 5 | EYA3 | 8936 2352 | 2294 | 1.122 | 0.0608 | No | ||
| 6 | SFPQ | 12936 | 2659 | 0.820 | 0.0575 | No | ||
| 7 | PBX3 | 9531 2714 2705 | 3147 | 0.471 | 0.0406 | No | ||
| 8 | SYNCRIP | 3078 3035 3107 | 3530 | 0.309 | 0.0261 | No | ||
| 9 | CSNK1E | 6570 2211 11332 | 3766 | 0.239 | 0.0182 | No | ||
| 10 | PHF15 | 20467 8050 | 3986 | 0.184 | 0.0101 | No | ||
| 11 | BMI1 | 4448 15113 | 4154 | 0.154 | 0.0041 | No | ||
| 12 | STK3 | 7084 | 4281 | 0.137 | 0.0001 | No | ||
| 13 | PCDHGB4 | 13534 | 4361 | 0.125 | -0.0017 | No | ||
| 14 | SON | 5473 1657 1684 | 4552 | 0.100 | -0.0100 | No | ||
| 15 | WNT11 | 5876 | 4738 | 0.084 | -0.0183 | No | ||
| 16 | KLC2 | 23973 3701 | 5140 | 0.061 | -0.0387 | No | ||
| 17 | MATR3 | 2043 5081 | 5186 | 0.059 | -0.0399 | No | ||
| 18 | RFX3 | 9722 5377 3712 3762 | 5240 | 0.057 | -0.0417 | No | ||
| 19 | HOXB8 | 9111 | 6077 | 0.031 | -0.0861 | No | ||
| 20 | SP3 | 5483 | 6951 | 0.015 | -0.1328 | No | ||
| 21 | KCNH2 | 16592 | 7060 | 0.013 | -0.1384 | No | ||
| 22 | AKAP10 | 20411 7145 | 7190 | 0.011 | -0.1451 | No | ||
| 23 | PHF8 | 6770 | 7545 | 0.006 | -0.1641 | No | ||
| 24 | TBC1D5 | 1570 22943 | 8551 | -0.007 | -0.2181 | No | ||
| 25 | CCNA2 | 15357 | 10583 | -0.038 | -0.3267 | No | ||
| 26 | PCDHGB5 | 13535 | 10720 | -0.041 | -0.3333 | No | ||
| 27 | BHLHB3 | 16935 | 11506 | -0.057 | -0.3744 | No | ||
| 28 | BRCA1 | 20213 | 11755 | -0.064 | -0.3865 | No | ||
| 29 | EN1 | 387 14155 | 12477 | -0.092 | -0.4235 | No | ||
| 30 | YARS | 16071 | 12610 | -0.098 | -0.4287 | No | ||
| 31 | HOXA10 | 17145 989 | 13155 | -0.129 | -0.4554 | No | ||
| 32 | RHOA | 8624 4409 4410 | 13287 | -0.138 | -0.4598 | No | ||
| 33 | RAD23A | 3878 5348 18813 9685 | 13379 | -0.145 | -0.4618 | No | ||
| 34 | LHX6 | 9275 2914 | 13474 | -0.153 | -0.4638 | No | ||
| 35 | PIGN | 13865 | 13717 | -0.176 | -0.4733 | No | ||
| 36 | CLK1 | 13951 4127 4529 8748 | 14063 | -0.214 | -0.4877 | No | ||
| 37 | HOXC10 | 22342 | 14164 | -0.228 | -0.4885 | No | ||
| 38 | CLSTN1 | 7241 | 14583 | -0.302 | -0.5051 | No | ||
| 39 | STAG2 | 5521 | 14608 | -0.308 | -0.5002 | No | ||
| 40 | PPP1R12A | 3354 19886 | 14818 | -0.361 | -0.5043 | No | ||
| 41 | NFIL3 | 21463 | 14913 | -0.388 | -0.5017 | No | ||
| 42 | ATF2 | 4418 2759 | 15372 | -0.574 | -0.5150 | No | ||
| 43 | UBE2D3 | 7253 | 15527 | -0.667 | -0.5100 | No | ||
| 44 | NR2E1 | 19780 | 16161 | -1.099 | -0.5223 | Yes | ||
| 45 | NKIRAS2 | 12979 | 16291 | -1.219 | -0.5051 | Yes | ||
| 46 | NFKBIA | 21065 | 16384 | -1.316 | -0.4839 | Yes | ||
| 47 | DHX15 | 8842 | 16474 | -1.430 | -0.4603 | Yes | ||
| 48 | SUCLG2 | 17062 5545 | 16480 | -1.436 | -0.4320 | Yes | ||
| 49 | YWHAE | 20776 | 16798 | -1.748 | -0.4144 | Yes | ||
| 50 | B4GALT1 | 15918 | 17104 | -2.180 | -0.3875 | Yes | ||
| 51 | PRPS1 | 24233 | 17165 | -2.238 | -0.3463 | Yes | ||
| 52 | ARL6IP2 | 1496 7090 12093 | 17225 | -2.344 | -0.3030 | Yes | ||
| 53 | ISL1 | 21338 | 17228 | -2.358 | -0.2563 | Yes | ||
| 54 | RBMX | 5367 9708 | 17257 | -2.417 | -0.2098 | Yes | ||
| 55 | DNAJC7 | 20227 | 17784 | -3.518 | -0.1682 | Yes | ||
| 56 | MIR16 | 17663 1354 | 18084 | -4.529 | -0.0944 | Yes | ||
| 57 | GART | 22543 1754 | 18431 | -6.196 | 0.0100 | Yes |