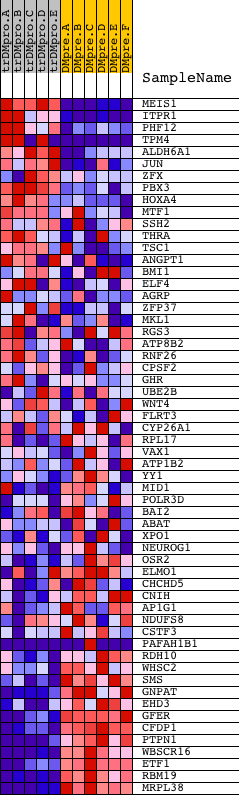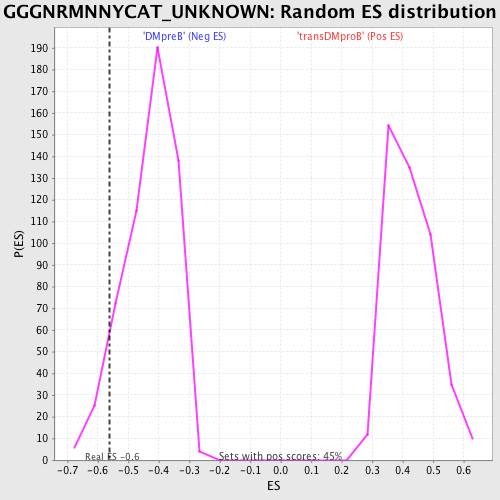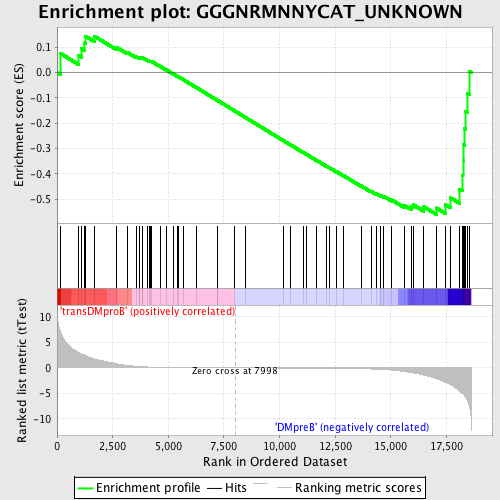
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | GGGNRMNNYCAT_UNKNOWN |
| Enrichment Score (ES) | -0.560908 |
| Normalized Enrichment Score (NES) | -1.3035793 |
| Nominal p-value | 0.07818182 |
| FDR q-value | 0.521252 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MEIS1 | 20524 | 167 | 6.817 | 0.0745 | No | ||
| 2 | ITPR1 | 17341 | 977 | 2.997 | 0.0676 | No | ||
| 3 | PHF12 | 11192 6483 | 1107 | 2.699 | 0.0937 | No | ||
| 4 | TPM4 | 11589 11588 6819 | 1238 | 2.444 | 0.1166 | No | ||
| 5 | ALDH6A1 | 21025 | 1280 | 2.351 | 0.1432 | No | ||
| 6 | JUN | 15832 | 1670 | 1.759 | 0.1438 | No | ||
| 7 | ZFX | 5984 | 2667 | 0.813 | 0.1001 | No | ||
| 8 | PBX3 | 9531 2714 2705 | 3147 | 0.471 | 0.0800 | No | ||
| 9 | HOXA4 | 17148 | 3560 | 0.297 | 0.0615 | No | ||
| 10 | MTF1 | 5128 9421 | 3694 | 0.257 | 0.0574 | No | ||
| 11 | SSH2 | 6202 | 3716 | 0.252 | 0.0594 | No | ||
| 12 | THRA | 1447 10171 1406 | 3826 | 0.224 | 0.0563 | No | ||
| 13 | TSC1 | 7235 | 3838 | 0.221 | 0.0584 | No | ||
| 14 | ANGPT1 | 8588 2260 22303 | 4073 | 0.167 | 0.0478 | No | ||
| 15 | BMI1 | 4448 15113 | 4154 | 0.154 | 0.0454 | No | ||
| 16 | ELF4 | 24162 | 4199 | 0.148 | 0.0448 | No | ||
| 17 | AGRP | 18768 | 4257 | 0.140 | 0.0435 | No | ||
| 18 | ZFP37 | 5962 | 4645 | 0.092 | 0.0238 | No | ||
| 19 | MKL1 | 5864 | 4933 | 0.072 | 0.0092 | No | ||
| 20 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 5251 | 0.056 | -0.0072 | No | ||
| 21 | ATP8B2 | 12055 | 5417 | 0.049 | -0.0155 | No | ||
| 22 | RNF26 | 5614 | 5462 | 0.047 | -0.0173 | No | ||
| 23 | CPSF2 | 21182 | 5662 | 0.041 | -0.0275 | No | ||
| 24 | GHR | 22336 | 6278 | 0.027 | -0.0603 | No | ||
| 25 | UBE2B | 10245 | 7199 | 0.011 | -0.1098 | No | ||
| 26 | WNT4 | 16025 | 7200 | 0.011 | -0.1096 | No | ||
| 27 | FLRT3 | 7730 12930 2673 | 7954 | 0.001 | -0.1502 | No | ||
| 28 | CYP26A1 | 23871 3695 | 8466 | -0.006 | -0.1777 | No | ||
| 29 | RPL17 | 11429 6653 | 10174 | -0.031 | -0.2692 | No | ||
| 30 | VAX1 | 23636 | 10511 | -0.037 | -0.2869 | No | ||
| 31 | ATP1B2 | 20390 | 11087 | -0.048 | -0.3173 | No | ||
| 32 | YY1 | 10371 | 11213 | -0.051 | -0.3234 | No | ||
| 33 | MID1 | 5097 5098 | 11671 | -0.062 | -0.3473 | No | ||
| 34 | POLR3D | 21760 12456 | 12108 | -0.076 | -0.3698 | No | ||
| 35 | BAI2 | 2541 2379 16067 | 12224 | -0.080 | -0.3750 | No | ||
| 36 | ABAT | 22864 1713 22669 | 12569 | -0.096 | -0.3924 | No | ||
| 37 | XPO1 | 4172 | 12880 | -0.112 | -0.4077 | No | ||
| 38 | NEUROG1 | 21446 | 13662 | -0.171 | -0.4477 | No | ||
| 39 | OSR2 | 22492 | 14112 | -0.221 | -0.4692 | No | ||
| 40 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 14344 | -0.253 | -0.4786 | No | ||
| 41 | CHCHD5 | 14856 | 14545 | -0.292 | -0.4858 | No | ||
| 42 | CNIH | 4536 23971 | 14675 | -0.325 | -0.4887 | No | ||
| 43 | AP1G1 | 4392 | 15035 | -0.425 | -0.5029 | No | ||
| 44 | NDUFS8 | 23950 | 15598 | -0.708 | -0.5245 | No | ||
| 45 | CSTF3 | 10449 6002 | 15917 | -0.952 | -0.5299 | No | ||
| 46 | PAFAH1B1 | 1340 5220 9524 | 15999 | -1.000 | -0.5221 | No | ||
| 47 | RDH10 | 8272 | 16486 | -1.440 | -0.5306 | No | ||
| 48 | WHSC2 | 6298 | 17049 | -2.098 | -0.5352 | Yes | ||
| 49 | SMS | 2573 24023 | 17445 | -2.797 | -0.5222 | Yes | ||
| 50 | GNPAT | 18420 | 17680 | -3.286 | -0.4946 | Yes | ||
| 51 | EHD3 | 23159 | 18083 | -4.526 | -0.4608 | Yes | ||
| 52 | GFER | 23088 | 18217 | -4.993 | -0.4069 | Yes | ||
| 53 | CFDP1 | 10710 | 18245 | -5.077 | -0.3462 | Yes | ||
| 54 | PTPN1 | 5325 | 18251 | -5.099 | -0.2840 | Yes | ||
| 55 | WBSCR16 | 16355 | 18305 | -5.354 | -0.2213 | Yes | ||
| 56 | ETF1 | 23467 | 18367 | -5.743 | -0.1542 | Yes | ||
| 57 | RBM19 | 13164 | 18428 | -6.177 | -0.0818 | Yes | ||
| 58 | MRPL38 | 20145 | 18541 | -7.505 | 0.0040 | Yes |