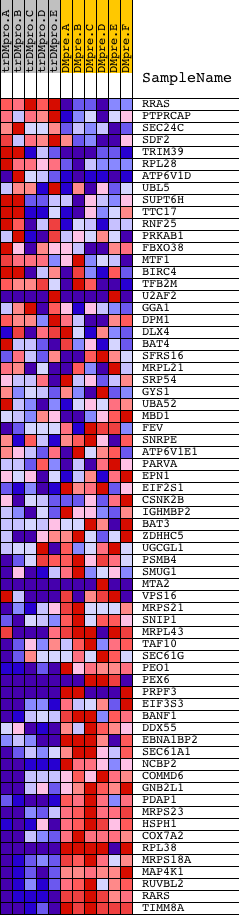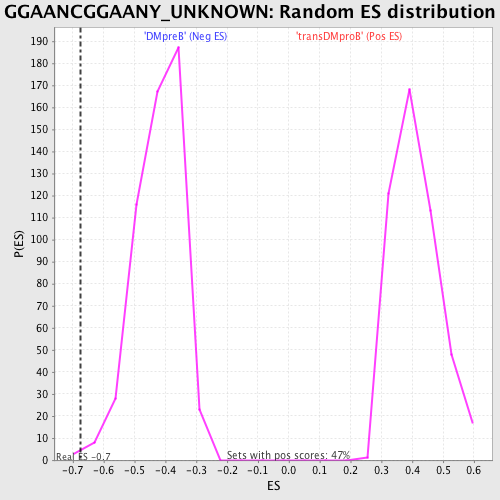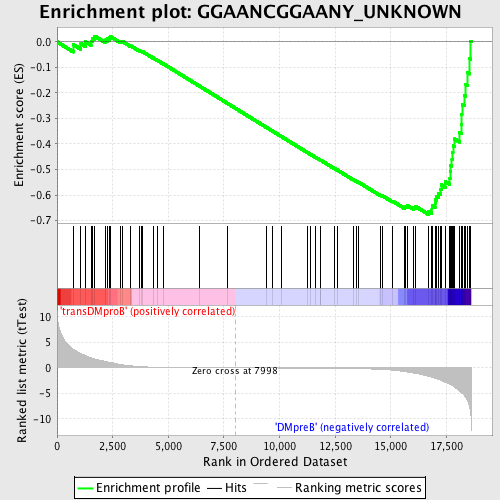
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | GGAANCGGAANY_UNKNOWN |
| Enrichment Score (ES) | -0.67633903 |
| Normalized Enrichment Score (NES) | -1.6103092 |
| Nominal p-value | 0.0037593986 |
| FDR q-value | 0.09180971 |
| FWER p-Value | 0.247 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RRAS | 18256 | 735 | 3.612 | -0.0110 | No | ||
| 2 | PTPRCAP | 23767 | 1069 | 2.788 | -0.0069 | No | ||
| 3 | SEC24C | 22086 | 1269 | 2.375 | 0.0012 | No | ||
| 4 | SDF2 | 20762 | 1526 | 1.955 | 0.0028 | No | ||
| 5 | TRIM39 | 22994 | 1585 | 1.878 | 0.0146 | No | ||
| 6 | RPL28 | 5392 | 1691 | 1.736 | 0.0226 | No | ||
| 7 | ATP6V1D | 7872 | 2157 | 1.261 | 0.0076 | No | ||
| 8 | UBL5 | 12299 | 2247 | 1.167 | 0.0120 | No | ||
| 9 | SUPT6H | 9940 | 2336 | 1.086 | 0.0158 | No | ||
| 10 | TTC17 | 13219 | 2394 | 1.037 | 0.0210 | No | ||
| 11 | RNF25 | 13922 3929 | 2861 | 0.644 | 0.0010 | No | ||
| 12 | PRKAB1 | 16408 | 2954 | 0.580 | 0.0006 | No | ||
| 13 | FBXO38 | 23421 | 3297 | 0.405 | -0.0146 | No | ||
| 14 | MTF1 | 5128 9421 | 3694 | 0.257 | -0.0340 | No | ||
| 15 | BIRC4 | 2638 8609 | 3793 | 0.231 | -0.0374 | No | ||
| 16 | TFB2M | 4024 9092 4083 | 3843 | 0.221 | -0.0383 | No | ||
| 17 | U2AF2 | 5813 5812 5814 | 4319 | 0.132 | -0.0629 | No | ||
| 18 | GGA1 | 22432 4197 | 4490 | 0.109 | -0.0712 | No | ||
| 19 | DPM1 | 4636 2882 8860 | 4775 | 0.081 | -0.0858 | No | ||
| 20 | DLX4 | 20282 1225 | 6380 | 0.025 | -0.1721 | No | ||
| 21 | BAT4 | 8172 8173 | 7671 | 0.004 | -0.2416 | No | ||
| 22 | SFRS16 | 17943 3840 | 9394 | -0.019 | -0.3343 | No | ||
| 23 | MRPL21 | 23755 | 9684 | -0.023 | -0.3497 | No | ||
| 24 | SRP54 | 6282 | 10085 | -0.030 | -0.3710 | No | ||
| 25 | GYS1 | 4818 | 11272 | -0.052 | -0.4346 | No | ||
| 26 | UBA52 | 10239 | 11370 | -0.054 | -0.4394 | No | ||
| 27 | MBD1 | 1959 23518 1998 2020 | 11605 | -0.060 | -0.4515 | No | ||
| 28 | FEV | 11153 6434 | 11830 | -0.067 | -0.4631 | No | ||
| 29 | SNRPE | 9843 | 11850 | -0.067 | -0.4635 | No | ||
| 30 | ATP6V1E1 | 4423 | 12449 | -0.091 | -0.4951 | No | ||
| 31 | PARVA | 18122 | 12581 | -0.097 | -0.5014 | No | ||
| 32 | EPN1 | 18401 | 13314 | -0.140 | -0.5397 | No | ||
| 33 | EIF2S1 | 4658 | 13442 | -0.150 | -0.5454 | No | ||
| 34 | CSNK2B | 23008 | 13539 | -0.159 | -0.5493 | No | ||
| 35 | IGHMBP2 | 23945 | 14542 | -0.290 | -0.6010 | No | ||
| 36 | BAT3 | 5896 | 14606 | -0.307 | -0.6020 | No | ||
| 37 | ZDHHC5 | 14547 | 15079 | -0.445 | -0.6239 | No | ||
| 38 | UGCGL1 | 6721 | 15631 | -0.730 | -0.6478 | No | ||
| 39 | PSMB4 | 15252 | 15677 | -0.765 | -0.6442 | No | ||
| 40 | SMUG1 | 22109 | 15743 | -0.818 | -0.6412 | No | ||
| 41 | MTA2 | 23934 18250 | 16021 | -1.007 | -0.6482 | No | ||
| 42 | VPS16 | 2927 14845 2825 | 16105 | -1.061 | -0.6443 | No | ||
| 43 | MRPS21 | 12323 | 16701 | -1.635 | -0.6634 | Yes | ||
| 44 | SNIP1 | 16085 | 16847 | -1.807 | -0.6569 | Yes | ||
| 45 | MRPL43 | 3703 23661 | 16850 | -1.815 | -0.6427 | Yes | ||
| 46 | TAF10 | 10785 | 16990 | -2.022 | -0.6341 | Yes | ||
| 47 | SEC61G | 9795 | 17000 | -2.030 | -0.6186 | Yes | ||
| 48 | PEO1 | 5932 | 17051 | -2.101 | -0.6046 | Yes | ||
| 49 | PEX6 | 5900 10331 | 17153 | -2.226 | -0.5925 | Yes | ||
| 50 | PRPF3 | 12899 12898 1877 7703 | 17238 | -2.379 | -0.5782 | Yes | ||
| 51 | EIF3S3 | 12652 | 17269 | -2.460 | -0.5603 | Yes | ||
| 52 | BANF1 | 6216 10706 3760 | 17446 | -2.797 | -0.5477 | Yes | ||
| 53 | DDX55 | 7490 | 17653 | -3.204 | -0.5334 | Yes | ||
| 54 | EBNA1BP2 | 8112 7599 12770 | 17665 | -3.236 | -0.5084 | Yes | ||
| 55 | SEC61A1 | 60 | 17683 | -3.289 | -0.4833 | Yes | ||
| 56 | NCBP2 | 12643 | 17747 | -3.425 | -0.4595 | Yes | ||
| 57 | COMMD6 | 476 | 17781 | -3.516 | -0.4335 | Yes | ||
| 58 | GNB2L1 | 20911 | 17795 | -3.543 | -0.4062 | Yes | ||
| 59 | PDAP1 | 16294 | 17866 | -3.741 | -0.3803 | Yes | ||
| 60 | MRPS23 | 12260 | 18094 | -4.580 | -0.3563 | Yes | ||
| 61 | HSPH1 | 16282 | 18166 | -4.828 | -0.3219 | Yes | ||
| 62 | COX7A2 | 19053 | 18175 | -4.873 | -0.2838 | Yes | ||
| 63 | RPL38 | 12562 20606 7475 | 18200 | -4.955 | -0.2459 | Yes | ||
| 64 | MRPS18A | 23222 | 18299 | -5.322 | -0.2090 | Yes | ||
| 65 | MAP4K1 | 18313 | 18371 | -5.767 | -0.1672 | Yes | ||
| 66 | RUVBL2 | 9766 | 18447 | -6.380 | -0.1207 | Yes | ||
| 67 | RARS | 20496 | 18543 | -7.573 | -0.0659 | Yes | ||
| 68 | TIMM8A | 24062 | 18588 | -8.817 | 0.0015 | Yes |