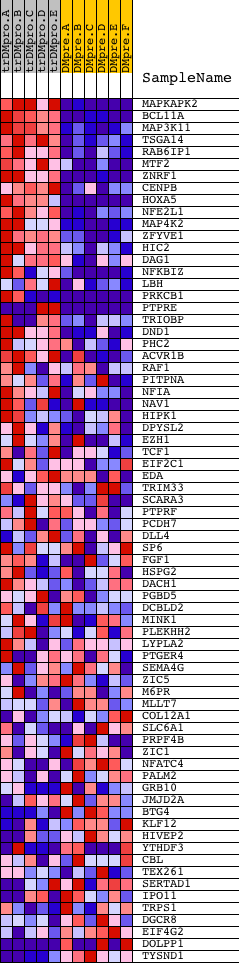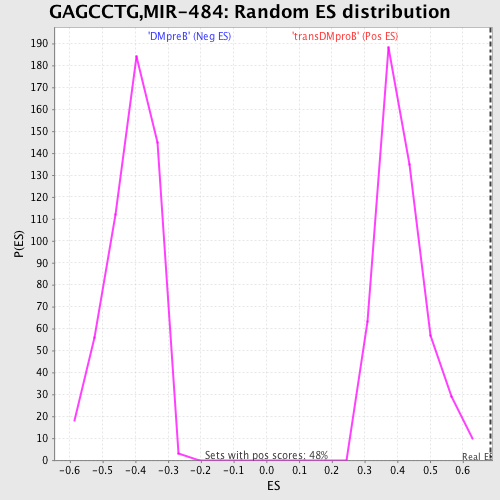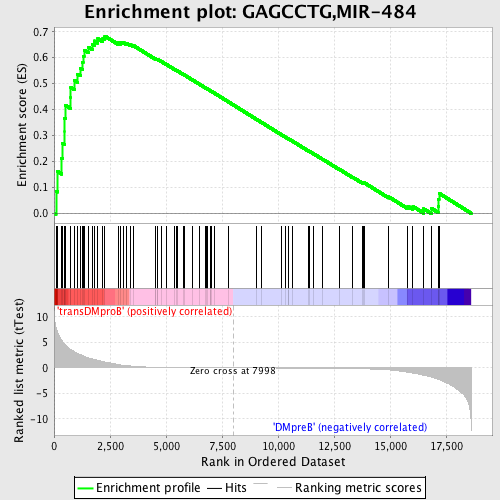
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | GAGCCTG,MIR-484 |
| Enrichment Score (ES) | 0.6832515 |
| Normalized Enrichment Score (NES) | 1.6500592 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.032822274 |
| FWER p-Value | 0.139 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MAPKAPK2 | 13838 | 85 | 7.980 | 0.0859 | Yes | ||
| 2 | BCL11A | 4691 | 159 | 6.951 | 0.1608 | Yes | ||
| 3 | MAP3K11 | 11163 | 339 | 5.379 | 0.2122 | Yes | ||
| 4 | TSGA14 | 17194 | 364 | 5.178 | 0.2696 | Yes | ||
| 5 | RAB6IP1 | 17676 | 473 | 4.566 | 0.3156 | Yes | ||
| 6 | MTF2 | 5129 | 475 | 4.562 | 0.3672 | Yes | ||
| 7 | ZNRF1 | 9306 | 498 | 4.498 | 0.4171 | Yes | ||
| 8 | CENPB | 14424 | 739 | 3.606 | 0.4450 | Yes | ||
| 9 | HOXA5 | 17149 | 746 | 3.576 | 0.4853 | Yes | ||
| 10 | NFE2L1 | 9457 | 913 | 3.149 | 0.5120 | Yes | ||
| 11 | MAP4K2 | 6457 | 1046 | 2.839 | 0.5371 | Yes | ||
| 12 | ZFYVE1 | 10138 | 1182 | 2.559 | 0.5588 | Yes | ||
| 13 | HIC2 | 7185 | 1276 | 2.357 | 0.5805 | Yes | ||
| 14 | DAG1 | 18996 8837 | 1293 | 2.333 | 0.6061 | Yes | ||
| 15 | NFKBIZ | 22575 | 1338 | 2.248 | 0.6293 | Yes | ||
| 16 | LBH | 23160 | 1529 | 1.952 | 0.6412 | Yes | ||
| 17 | PRKCB1 | 1693 9574 | 1709 | 1.712 | 0.6509 | Yes | ||
| 18 | PTPRE | 1662 18039 | 1801 | 1.630 | 0.6645 | Yes | ||
| 19 | TRIOBP | 2280 670 8476 | 1953 | 1.455 | 0.6729 | Yes | ||
| 20 | DND1 | 10017 | 2177 | 1.242 | 0.6749 | Yes | ||
| 21 | PHC2 | 16075 | 2265 | 1.148 | 0.6833 | Yes | ||
| 22 | ACVR1B | 4335 22353 | 2853 | 0.647 | 0.6589 | No | ||
| 23 | RAF1 | 17035 | 2978 | 0.564 | 0.6587 | No | ||
| 24 | PITPNA | 20778 | 3079 | 0.512 | 0.6591 | No | ||
| 25 | NFIA | 16172 5170 | 3241 | 0.430 | 0.6553 | No | ||
| 26 | NAV1 | 3978 5681 | 3402 | 0.360 | 0.6507 | No | ||
| 27 | HIPK1 | 4851 | 3522 | 0.312 | 0.6478 | No | ||
| 28 | DPYSL2 | 3122 4561 8787 | 4507 | 0.106 | 0.5960 | No | ||
| 29 | EZH1 | 20217 | 4546 | 0.101 | 0.5951 | No | ||
| 30 | TCF1 | 16416 | 4599 | 0.096 | 0.5934 | No | ||
| 31 | EIF2C1 | 10672 | 4632 | 0.093 | 0.5927 | No | ||
| 32 | EDA | 24284 | 4793 | 0.079 | 0.5850 | No | ||
| 33 | TRIM33 | 8236 13579 15472 8237 | 4997 | 0.068 | 0.5748 | No | ||
| 34 | SCARA3 | 21780 | 5394 | 0.050 | 0.5540 | No | ||
| 35 | PTPRF | 5331 2545 | 5442 | 0.048 | 0.5520 | No | ||
| 36 | PCDH7 | 7029 3460 | 5487 | 0.046 | 0.5502 | No | ||
| 37 | DLL4 | 7040 | 5785 | 0.038 | 0.5346 | No | ||
| 38 | SP6 | 8178 13678 13679 | 5835 | 0.037 | 0.5324 | No | ||
| 39 | FGF1 | 1994 23447 | 6173 | 0.029 | 0.5145 | No | ||
| 40 | HSPG2 | 4884 16023 | 6495 | 0.023 | 0.4975 | No | ||
| 41 | DACH1 | 4595 3371 | 6771 | 0.018 | 0.4828 | No | ||
| 42 | PGBD5 | 9960 | 6801 | 0.017 | 0.4815 | No | ||
| 43 | DCBLD2 | 7855 | 6836 | 0.016 | 0.4798 | No | ||
| 44 | MINK1 | 20802 | 6976 | 0.014 | 0.4725 | No | ||
| 45 | PLEKHH2 | 23148 | 6993 | 0.014 | 0.4718 | No | ||
| 46 | LYPLA2 | 15708 | 7038 | 0.013 | 0.4696 | No | ||
| 47 | PTGER4 | 9649 | 7170 | 0.011 | 0.4626 | No | ||
| 48 | SEMA4G | 11182 | 7790 | 0.003 | 0.4293 | No | ||
| 49 | ZIC5 | 12268 | 9023 | -0.014 | 0.3630 | No | ||
| 50 | M6PR | 1053 5044 9328 | 9240 | -0.017 | 0.3515 | No | ||
| 51 | MLLT7 | 24278 | 10147 | -0.031 | 0.3030 | No | ||
| 52 | COL12A1 | 3143 19054 | 10329 | -0.034 | 0.2937 | No | ||
| 53 | SLC6A1 | 10557 | 10451 | -0.036 | 0.2876 | No | ||
| 54 | PRPF4B | 3208 5295 | 10475 | -0.036 | 0.2867 | No | ||
| 55 | ZIC1 | 19040 | 10649 | -0.039 | 0.2779 | No | ||
| 56 | NFATC4 | 22002 | 11337 | -0.054 | 0.2414 | No | ||
| 57 | PALM2 | 10817 2368 | 11410 | -0.055 | 0.2382 | No | ||
| 58 | GRB10 | 4799 | 11590 | -0.060 | 0.2292 | No | ||
| 59 | JMJD2A | 10505 6058 | 11981 | -0.072 | 0.2090 | No | ||
| 60 | BTG4 | 19456 | 12754 | -0.105 | 0.1685 | No | ||
| 61 | KLF12 | 4960 2637 21732 9228 | 13321 | -0.141 | 0.1396 | No | ||
| 62 | HIVEP2 | 9091 | 13770 | -0.182 | 0.1175 | No | ||
| 63 | YTHDF3 | 15630 10469 6019 | 13802 | -0.185 | 0.1179 | No | ||
| 64 | CBL | 19154 | 13840 | -0.189 | 0.1181 | No | ||
| 65 | TEX261 | 17094 5726 | 14937 | -0.396 | 0.0635 | No | ||
| 66 | SERTAD1 | 18326 | 15794 | -0.854 | 0.0270 | No | ||
| 67 | IPO11 | 21353 | 16019 | -1.005 | 0.0263 | No | ||
| 68 | TRPS1 | 8195 | 16501 | -1.463 | 0.0170 | No | ||
| 69 | DGCR8 | 22644 | 16851 | -1.817 | 0.0188 | No | ||
| 70 | EIF4G2 | 1908 8892 | 17147 | -2.222 | 0.0281 | No | ||
| 71 | DOLPP1 | 2682 15051 | 17162 | -2.236 | 0.0527 | No | ||
| 72 | TYSND1 | 20007 | 17183 | -2.265 | 0.0773 | No |