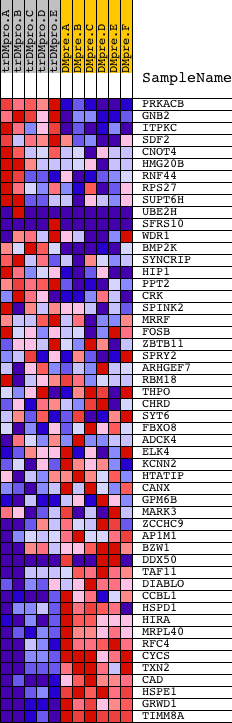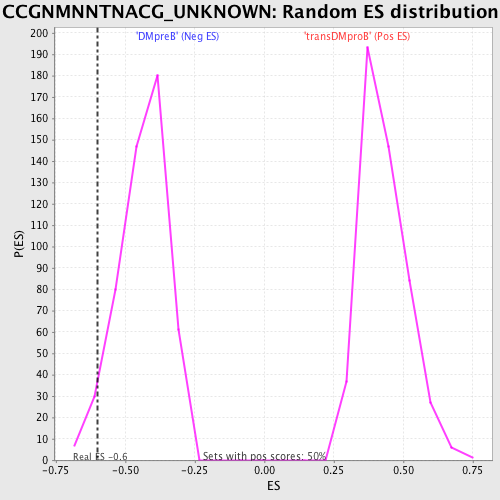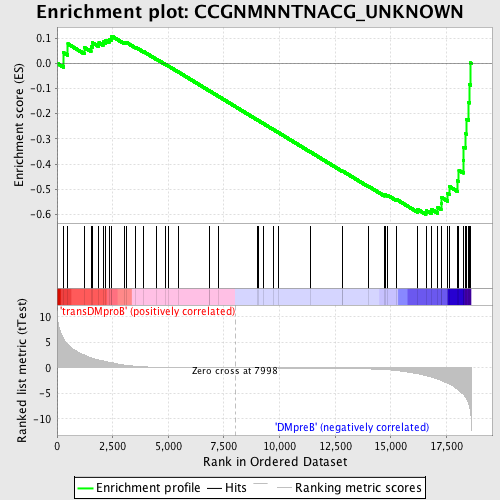
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | CCGNMNNTNACG_UNKNOWN |
| Enrichment Score (ES) | -0.5997622 |
| Normalized Enrichment Score (NES) | -1.3729912 |
| Nominal p-value | 0.03960396 |
| FDR q-value | 0.41540307 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PRKACB | 15140 | 286 | 5.707 | 0.0421 | No | ||
| 2 | GNB2 | 9026 | 487 | 4.520 | 0.0769 | No | ||
| 3 | ITPKC | 17919 | 1220 | 2.495 | 0.0626 | No | ||
| 4 | SDF2 | 20762 | 1526 | 1.955 | 0.0659 | No | ||
| 5 | CNOT4 | 7010 1022 | 1581 | 1.884 | 0.0819 | No | ||
| 6 | HMG20B | 19676 | 1880 | 1.536 | 0.0814 | No | ||
| 7 | RNF44 | 8364 | 2067 | 1.340 | 0.0848 | No | ||
| 8 | RPS27 | 12183 | 2173 | 1.245 | 0.0917 | No | ||
| 9 | SUPT6H | 9940 | 2336 | 1.086 | 0.0939 | No | ||
| 10 | UBE2H | 5823 5822 | 2433 | 1.005 | 0.0989 | No | ||
| 11 | SFRS10 | 9818 1722 5441 | 2457 | 0.997 | 0.1077 | No | ||
| 12 | WDR1 | 16544 3592 | 3016 | 0.547 | 0.0832 | No | ||
| 13 | BMP2K | 3657 16786 | 3139 | 0.478 | 0.0814 | No | ||
| 14 | SYNCRIP | 3078 3035 3107 | 3530 | 0.309 | 0.0635 | No | ||
| 15 | HIP1 | 5676 | 3903 | 0.203 | 0.0455 | No | ||
| 16 | PPT2 | 1617 12034 | 4459 | 0.112 | 0.0168 | No | ||
| 17 | CRK | 4559 1249 | 4878 | 0.075 | -0.0050 | No | ||
| 18 | SPINK2 | 16502 | 5012 | 0.068 | -0.0115 | No | ||
| 19 | MRRF | 12594 2837 | 5467 | 0.047 | -0.0355 | No | ||
| 20 | FOSB | 17945 | 6834 | 0.016 | -0.1089 | No | ||
| 21 | ZBTB11 | 11309 | 6853 | 0.016 | -0.1097 | No | ||
| 22 | SPRY2 | 21725 | 7274 | 0.010 | -0.1322 | No | ||
| 23 | ARHGEF7 | 3823 | 8994 | -0.014 | -0.2247 | No | ||
| 24 | RBM18 | 2789 14604 | 9066 | -0.015 | -0.2283 | No | ||
| 25 | THPO | 22636 | 9280 | -0.018 | -0.2396 | No | ||
| 26 | CHRD | 22817 | 9736 | -0.024 | -0.2639 | No | ||
| 27 | SYT6 | 13437 7042 12040 | 9929 | -0.027 | -0.2740 | No | ||
| 28 | FBXO8 | 18616 3896 | 11369 | -0.054 | -0.3509 | No | ||
| 29 | ADCK4 | 13364 | 12806 | -0.108 | -0.4272 | No | ||
| 30 | ELK4 | 8895 4664 3937 | 12844 | -0.110 | -0.4281 | No | ||
| 31 | KCNN2 | 8933 1949 | 14000 | -0.207 | -0.4882 | No | ||
| 32 | HTATIP | 3690 | 14713 | -0.337 | -0.5232 | No | ||
| 33 | CANX | 4474 | 14747 | -0.345 | -0.5215 | No | ||
| 34 | GPM6B | 9034 | 14849 | -0.369 | -0.5232 | No | ||
| 35 | MARK3 | 9369 | 15234 | -0.507 | -0.5387 | No | ||
| 36 | ZCCHC9 | 21398 | 16201 | -1.140 | -0.5793 | No | ||
| 37 | AP1M1 | 18571 | 16582 | -1.540 | -0.5842 | Yes | ||
| 38 | BZW1 | 7356 | 16829 | -1.789 | -0.5795 | Yes | ||
| 39 | DDX50 | 3324 19749 | 17112 | -2.188 | -0.5726 | Yes | ||
| 40 | TAF11 | 12733 | 17261 | -2.429 | -0.5561 | Yes | ||
| 41 | DIABLO | 16375 | 17272 | -2.465 | -0.5318 | Yes | ||
| 42 | CCBL1 | 14630 | 17552 | -2.995 | -0.5166 | Yes | ||
| 43 | HSPD1 | 4078 9129 | 17647 | -3.193 | -0.4895 | Yes | ||
| 44 | HIRA | 4852 9090 | 18002 | -4.201 | -0.4663 | Yes | ||
| 45 | MRPL40 | 22641 | 18062 | -4.439 | -0.4247 | Yes | ||
| 46 | RFC4 | 1735 22627 | 18260 | -5.128 | -0.3837 | Yes | ||
| 47 | CYCS | 8821 | 18278 | -5.207 | -0.3321 | Yes | ||
| 48 | TXN2 | 22227 | 18356 | -5.657 | -0.2792 | Yes | ||
| 49 | CAD | 16886 | 18408 | -6.035 | -0.2212 | Yes | ||
| 50 | HSPE1 | 9133 | 18500 | -6.948 | -0.1561 | Yes | ||
| 51 | GRWD1 | 8317 | 18531 | -7.280 | -0.0843 | Yes | ||
| 52 | TIMM8A | 24062 | 18588 | -8.817 | 0.0015 | Yes |