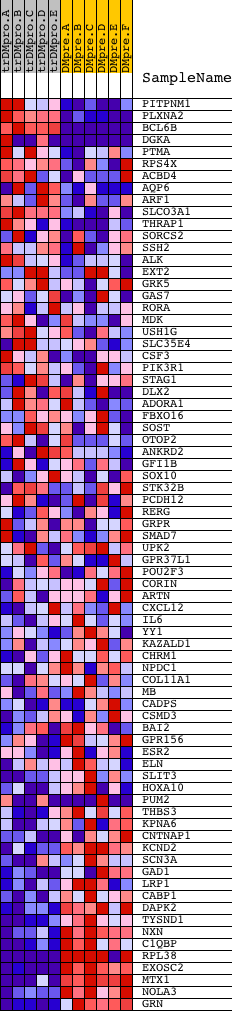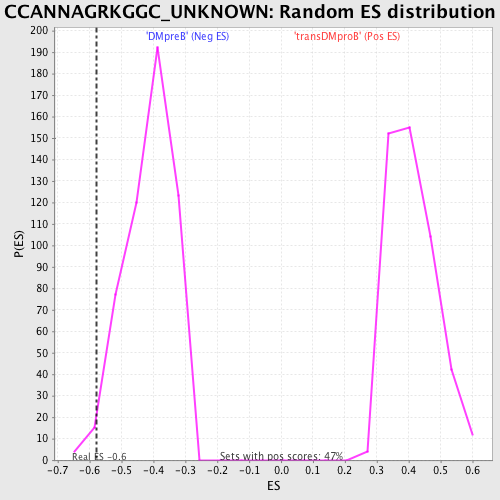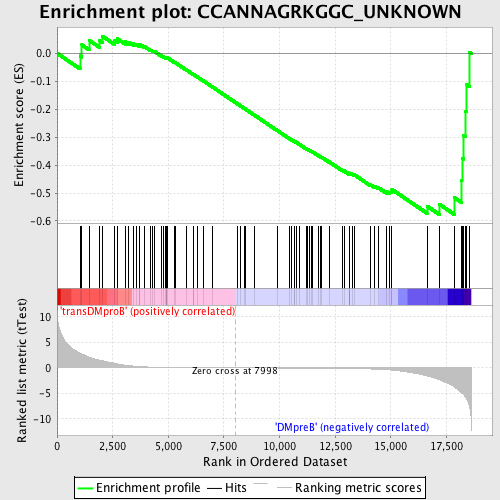
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | CCANNAGRKGGC_UNKNOWN |
| Enrichment Score (ES) | -0.5768891 |
| Normalized Enrichment Score (NES) | -1.392297 |
| Nominal p-value | 0.02636535 |
| FDR q-value | 0.40143955 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PITPNM1 | 23770 | 1033 | 2.865 | -0.0093 | No | ||
| 2 | PLXNA2 | 5269 | 1114 | 2.689 | 0.0300 | No | ||
| 3 | BCL6B | 20373 | 1437 | 2.084 | 0.0464 | No | ||
| 4 | DGKA | 3359 19589 | 1891 | 1.523 | 0.0467 | No | ||
| 5 | PTMA | 9657 | 2030 | 1.375 | 0.0615 | No | ||
| 6 | RPS4X | 9756 | 2598 | 0.876 | 0.0451 | No | ||
| 7 | ACBD4 | 20637 | 2704 | 0.779 | 0.0521 | No | ||
| 8 | AQP6 | 22364 8621 | 3060 | 0.520 | 0.0414 | No | ||
| 9 | ARF1 | 64 | 3224 | 0.439 | 0.0397 | No | ||
| 10 | SLCO3A1 | 17796 4237 8430 | 3423 | 0.351 | 0.0347 | No | ||
| 11 | THRAP1 | 20309 | 3548 | 0.304 | 0.0330 | No | ||
| 12 | SORCS2 | 16553 | 3704 | 0.256 | 0.0288 | No | ||
| 13 | SSH2 | 6202 | 3716 | 0.252 | 0.0323 | No | ||
| 14 | ALK | 8574 | 3947 | 0.192 | 0.0230 | No | ||
| 15 | EXT2 | 14516 | 4214 | 0.145 | 0.0110 | No | ||
| 16 | GRK5 | 9036 23814 | 4307 | 0.133 | 0.0082 | No | ||
| 17 | GAS7 | 20836 1299 | 4366 | 0.123 | 0.0070 | No | ||
| 18 | RORA | 3019 5388 | 4686 | 0.088 | -0.0087 | No | ||
| 19 | MDK | 14523 | 4786 | 0.080 | -0.0128 | No | ||
| 20 | USH1G | 20153 | 4891 | 0.074 | -0.0172 | No | ||
| 21 | SLC35E4 | 20549 | 4905 | 0.073 | -0.0167 | No | ||
| 22 | CSF3 | 1394 20671 | 4912 | 0.073 | -0.0159 | No | ||
| 23 | PIK3R1 | 3170 | 4927 | 0.072 | -0.0154 | No | ||
| 24 | STAG1 | 5520 9905 2987 | 4970 | 0.069 | -0.0166 | No | ||
| 25 | DLX2 | 14561 353 | 5282 | 0.055 | -0.0325 | No | ||
| 26 | ADORA1 | 13831 | 5319 | 0.053 | -0.0335 | No | ||
| 27 | FBXO16 | 11929 | 5797 | 0.038 | -0.0587 | No | ||
| 28 | SOST | 20210 | 6128 | 0.030 | -0.0760 | No | ||
| 29 | OTOP2 | 20600 | 6140 | 0.030 | -0.0761 | No | ||
| 30 | ANKRD2 | 23853 | 6320 | 0.026 | -0.0853 | No | ||
| 31 | GFI1B | 14637 | 6589 | 0.021 | -0.0994 | No | ||
| 32 | SOX10 | 22211 | 6990 | 0.014 | -0.1208 | No | ||
| 33 | STK32B | 12255 | 8085 | -0.001 | -0.1798 | No | ||
| 34 | PCDH12 | 7005 | 8253 | -0.003 | -0.1887 | No | ||
| 35 | RERG | 16949 | 8430 | -0.006 | -0.1981 | No | ||
| 36 | GRPR | 24015 | 8457 | -0.006 | -0.1994 | No | ||
| 37 | SMAD7 | 23513 | 8883 | -0.012 | -0.2222 | No | ||
| 38 | UPK2 | 19146 | 9914 | -0.027 | -0.2773 | No | ||
| 39 | GPR37L1 | 13825 | 10464 | -0.036 | -0.3063 | No | ||
| 40 | POU2F3 | 9602 | 10537 | -0.038 | -0.3096 | No | ||
| 41 | CORIN | 16517 | 10673 | -0.040 | -0.3162 | No | ||
| 42 | ARTN | 15783 2430 2532 | 10683 | -0.040 | -0.3160 | No | ||
| 43 | CXCL12 | 9792 1182 1031 | 10763 | -0.041 | -0.3196 | No | ||
| 44 | IL6 | 16895 | 10906 | -0.045 | -0.3266 | No | ||
| 45 | YY1 | 10371 | 11213 | -0.051 | -0.3422 | No | ||
| 46 | KAZALD1 | 4206 | 11243 | -0.051 | -0.3430 | No | ||
| 47 | CHRM1 | 23943 | 11350 | -0.054 | -0.3478 | No | ||
| 48 | NPDC1 | 9479 | 11450 | -0.056 | -0.3522 | No | ||
| 49 | COL11A1 | 15442 1937 4540 8762 | 11480 | -0.057 | -0.3529 | No | ||
| 50 | MB | 22232 | 11756 | -0.064 | -0.3667 | No | ||
| 51 | CADPS | 11298 | 11853 | -0.067 | -0.3708 | No | ||
| 52 | CSMD3 | 10735 7938 | 11893 | -0.069 | -0.3718 | No | ||
| 53 | BAI2 | 2541 2379 16067 | 12224 | -0.080 | -0.3883 | No | ||
| 54 | GPR156 | 10753 22763 | 12824 | -0.109 | -0.4188 | No | ||
| 55 | ESR2 | 21038 | 12895 | -0.112 | -0.4208 | No | ||
| 56 | ELN | 8896 | 13133 | -0.127 | -0.4315 | No | ||
| 57 | SLIT3 | 20925 | 13152 | -0.129 | -0.4304 | No | ||
| 58 | HOXA10 | 17145 989 | 13155 | -0.129 | -0.4284 | No | ||
| 59 | PUM2 | 2071 8161 | 13266 | -0.137 | -0.4321 | No | ||
| 60 | THBS3 | 15542 1796 | 13345 | -0.142 | -0.4340 | No | ||
| 61 | KPNA6 | 4972 | 14098 | -0.219 | -0.4710 | No | ||
| 62 | CNTNAP1 | 11987 | 14259 | -0.242 | -0.4757 | No | ||
| 63 | KCND2 | 4941 17515 | 14438 | -0.271 | -0.4809 | No | ||
| 64 | SCN3A | 14573 | 14820 | -0.362 | -0.4956 | No | ||
| 65 | GAD1 | 14993 2690 | 14959 | -0.403 | -0.4965 | No | ||
| 66 | LRP1 | 9284 | 15017 | -0.419 | -0.4928 | No | ||
| 67 | CABP1 | 16412 | 15050 | -0.433 | -0.4875 | No | ||
| 68 | DAPK2 | 19395 | 16630 | -1.575 | -0.5471 | Yes | ||
| 69 | TYSND1 | 20007 | 17183 | -2.265 | -0.5402 | Yes | ||
| 70 | NXN | 5204 | 17849 | -3.705 | -0.5160 | Yes | ||
| 71 | C1QBP | 20364 | 18178 | -4.891 | -0.4544 | Yes | ||
| 72 | RPL38 | 12562 20606 7475 | 18200 | -4.955 | -0.3752 | Yes | ||
| 73 | EXOSC2 | 15044 | 18279 | -5.220 | -0.2947 | Yes | ||
| 74 | MTX1 | 15282 1918 | 18360 | -5.705 | -0.2066 | Yes | ||
| 75 | NOLA3 | 14914 | 18401 | -6.008 | -0.1113 | Yes | ||
| 76 | GRN | 20640 | 18544 | -7.579 | 0.0039 | Yes |