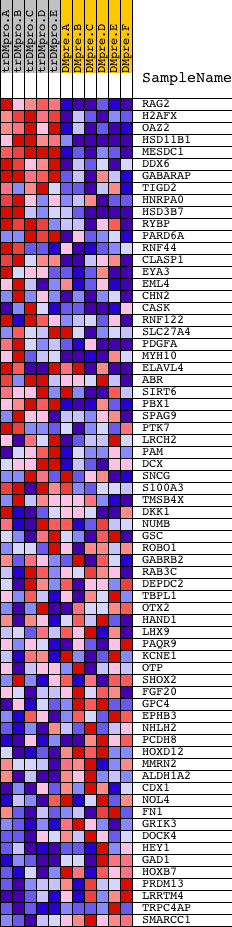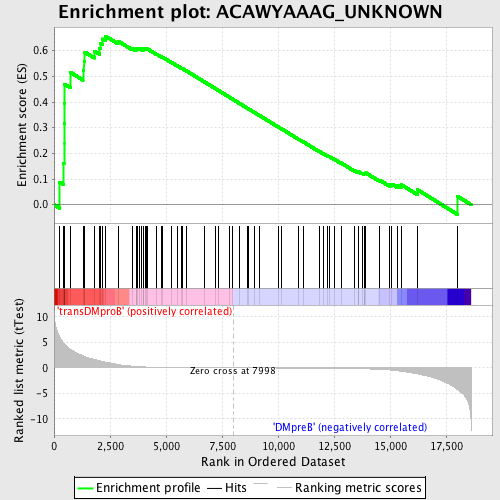
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | ACAWYAAAG_UNKNOWN |
| Enrichment Score (ES) | 0.6567473 |
| Normalized Enrichment Score (NES) | 1.600951 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.05791775 |
| FWER p-Value | 0.323 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RAG2 | 14941 | 247 | 6.107 | 0.0884 | Yes | ||
| 2 | H2AFX | 19477 | 415 | 4.883 | 0.1608 | Yes | ||
| 3 | OAZ2 | 9499 | 444 | 4.707 | 0.2376 | Yes | ||
| 4 | HSD11B1 | 9125 4019 | 446 | 4.702 | 0.3159 | Yes | ||
| 5 | MESDC1 | 17771 | 457 | 4.654 | 0.3929 | Yes | ||
| 6 | DDX6 | 3086 8844 | 465 | 4.606 | 0.4692 | Yes | ||
| 7 | GABARAP | 20815 | 725 | 3.636 | 0.5159 | Yes | ||
| 8 | TIGD2 | 17429 | 1290 | 2.337 | 0.5244 | Yes | ||
| 9 | HNRPA0 | 13382 | 1334 | 2.257 | 0.5596 | Yes | ||
| 10 | HSD3B7 | 4158 | 1374 | 2.189 | 0.5940 | Yes | ||
| 11 | RYBP | 17056 | 1794 | 1.633 | 0.5986 | Yes | ||
| 12 | PARD6A | 12142 | 2036 | 1.370 | 0.6084 | Yes | ||
| 13 | RNF44 | 8364 | 2067 | 1.340 | 0.6292 | Yes | ||
| 14 | CLASP1 | 14159 | 2154 | 1.262 | 0.6455 | Yes | ||
| 15 | EYA3 | 8936 2352 | 2294 | 1.122 | 0.6567 | Yes | ||
| 16 | EML4 | 8122 | 2858 | 0.646 | 0.6372 | No | ||
| 17 | CHN2 | 7652 | 3493 | 0.323 | 0.6084 | No | ||
| 18 | CASK | 24184 2618 2604 | 3660 | 0.267 | 0.6038 | No | ||
| 19 | RNF122 | 18639 | 3671 | 0.264 | 0.6077 | No | ||
| 20 | SLC27A4 | 15060 | 3717 | 0.252 | 0.6095 | No | ||
| 21 | PDGFA | 5237 | 3795 | 0.231 | 0.6092 | No | ||
| 22 | MYH10 | 8077 | 3920 | 0.200 | 0.6058 | No | ||
| 23 | ELAVL4 | 15805 4889 9137 | 3984 | 0.185 | 0.6055 | No | ||
| 24 | ABR | 8468 20346 1476 1429 | 4004 | 0.181 | 0.6075 | No | ||
| 25 | SIRT6 | 19668 3447 | 4080 | 0.166 | 0.6062 | No | ||
| 26 | PBX1 | 5224 | 4121 | 0.161 | 0.6067 | No | ||
| 27 | SPAG9 | 1322 12904 7707 | 4166 | 0.152 | 0.6069 | No | ||
| 28 | PTK7 | 22959 | 4569 | 0.098 | 0.5868 | No | ||
| 29 | LRCH2 | 24035 | 4787 | 0.080 | 0.5765 | No | ||
| 30 | PAM | 5222 | 4843 | 0.076 | 0.5748 | No | ||
| 31 | DCX | 24040 8841 4604 | 5248 | 0.056 | 0.5539 | No | ||
| 32 | SNCG | 21878 | 5511 | 0.046 | 0.5406 | No | ||
| 33 | S100A3 | 9772 | 5687 | 0.041 | 0.5318 | No | ||
| 34 | TMSB4X | 24005 | 5727 | 0.040 | 0.5304 | No | ||
| 35 | DKK1 | 23700 | 5901 | 0.035 | 0.5216 | No | ||
| 36 | NUMB | 5202 2146 2054 | 6731 | 0.018 | 0.4772 | No | ||
| 37 | GSC | 20990 | 7204 | 0.011 | 0.4520 | No | ||
| 38 | ROBO1 | 9731 | 7348 | 0.009 | 0.4444 | No | ||
| 39 | GABRB2 | 20922 | 7850 | 0.002 | 0.4174 | No | ||
| 40 | RAB3C | 21351 | 7947 | 0.001 | 0.4123 | No | ||
| 41 | DEPDC2 | 14295 | 8274 | -0.004 | 0.3947 | No | ||
| 42 | TBPL1 | 19805 | 8635 | -0.008 | 0.3755 | No | ||
| 43 | OTX2 | 9519 | 8660 | -0.009 | 0.3743 | No | ||
| 44 | HAND1 | 20440 | 8948 | -0.013 | 0.3591 | No | ||
| 45 | LHX9 | 9276 | 9156 | -0.016 | 0.3482 | No | ||
| 46 | PAQR9 | 19351 | 10003 | -0.029 | 0.3030 | No | ||
| 47 | KCNE1 | 9206 22537 4942 | 10164 | -0.031 | 0.2949 | No | ||
| 48 | OTP | 21582 | 10926 | -0.045 | 0.2546 | No | ||
| 49 | SHOX2 | 1802 5432 | 11113 | -0.049 | 0.2454 | No | ||
| 50 | FGF20 | 18892 | 11835 | -0.067 | 0.2077 | No | ||
| 51 | GPC4 | 24153 | 12034 | -0.073 | 0.1982 | No | ||
| 52 | EPHB3 | 22816 | 12211 | -0.080 | 0.1901 | No | ||
| 53 | NHLH2 | 9462 | 12288 | -0.084 | 0.1874 | No | ||
| 54 | PCDH8 | 21739 3017 | 12521 | -0.094 | 0.1764 | No | ||
| 55 | HOXD12 | 9113 | 12809 | -0.108 | 0.1627 | No | ||
| 56 | MMRN2 | 4194 | 13417 | -0.147 | 0.1325 | No | ||
| 57 | ALDH1A2 | 19386 | 13570 | -0.162 | 0.1270 | No | ||
| 58 | CDX1 | 23429 | 13604 | -0.165 | 0.1280 | No | ||
| 59 | NOL4 | 2028 11432 | 13758 | -0.181 | 0.1227 | No | ||
| 60 | FN1 | 4091 4094 971 13929 | 13834 | -0.188 | 0.1218 | No | ||
| 61 | GRIK3 | 16083 | 13875 | -0.193 | 0.1229 | No | ||
| 62 | DOCK4 | 10705 | 13920 | -0.197 | 0.1238 | No | ||
| 63 | HEY1 | 15378 | 14532 | -0.290 | 0.0957 | No | ||
| 64 | GAD1 | 14993 2690 | 14959 | -0.403 | 0.0794 | No | ||
| 65 | HOXB7 | 666 9110 | 15082 | -0.446 | 0.0803 | No | ||
| 66 | PRDM13 | 15933 | 15333 | -0.551 | 0.0760 | No | ||
| 67 | LRRTM4 | 17405 | 15489 | -0.648 | 0.0784 | No | ||
| 68 | TRPC4AP | 14379 12125 | 16230 | -1.167 | 0.0579 | No | ||
| 69 | SMARCC1 | 9834 5459 | 18012 | -4.240 | 0.0326 | No |