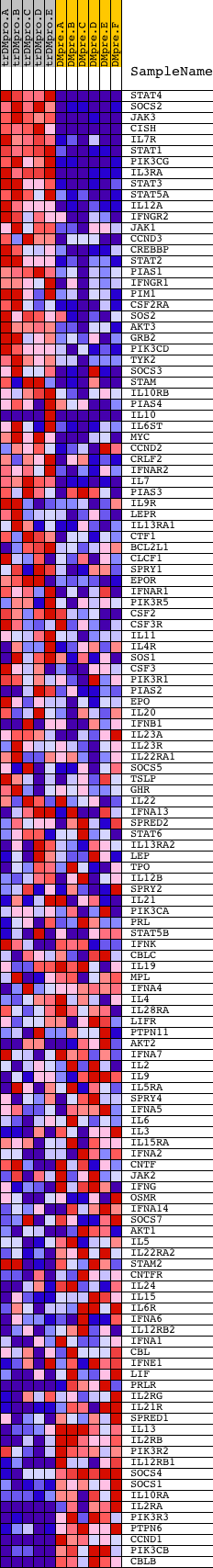
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | HSA04630_JAK_STAT_SIGNALING_PATHWAY |
| Enrichment Score (ES) | 0.5866086 |
| Normalized Enrichment Score (NES) | 1.5407474 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.15099296 |
| FWER p-Value | 0.891 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | STAT4 | 14251 9907 | 25 | 9.695 | 0.0626 | Yes | ||
| 2 | SOCS2 | 5694 | 65 | 8.301 | 0.1153 | Yes | ||
| 3 | JAK3 | 9198 4936 | 107 | 7.705 | 0.1639 | Yes | ||
| 4 | CISH | 8743 | 157 | 6.972 | 0.2073 | Yes | ||
| 5 | IL7R | 9175 4922 | 170 | 6.753 | 0.2512 | Yes | ||
| 6 | STAT1 | 3936 5524 | 190 | 6.574 | 0.2935 | Yes | ||
| 7 | PIK3CG | 6635 | 265 | 5.945 | 0.3288 | Yes | ||
| 8 | IL3RA | 22091 | 356 | 5.225 | 0.3584 | Yes | ||
| 9 | STAT3 | 5525 9906 | 569 | 4.247 | 0.3749 | Yes | ||
| 10 | STAT5A | 20664 | 727 | 3.634 | 0.3904 | Yes | ||
| 11 | IL12A | 4913 | 820 | 3.377 | 0.4077 | Yes | ||
| 12 | IFNGR2 | 9155 22702 | 877 | 3.244 | 0.4261 | Yes | ||
| 13 | JAK1 | 15827 | 996 | 2.955 | 0.4392 | Yes | ||
| 14 | CCND3 | 4489 4490 | 1057 | 2.813 | 0.4545 | Yes | ||
| 15 | CREBBP | 22682 8783 | 1098 | 2.720 | 0.4703 | Yes | ||
| 16 | STAT2 | 19840 | 1162 | 2.593 | 0.4840 | Yes | ||
| 17 | PIAS1 | 7126 | 1179 | 2.563 | 0.5000 | Yes | ||
| 18 | IFNGR1 | 20083 | 1251 | 2.410 | 0.5121 | Yes | ||
| 19 | PIM1 | 23308 | 1381 | 2.178 | 0.5195 | Yes | ||
| 20 | CSF2RA | 23631 | 1422 | 2.114 | 0.5313 | Yes | ||
| 21 | SOS2 | 21049 | 1479 | 2.025 | 0.5416 | Yes | ||
| 22 | AKT3 | 13739 982 | 1498 | 1.998 | 0.5538 | Yes | ||
| 23 | GRB2 | 20149 | 1534 | 1.943 | 0.5648 | Yes | ||
| 24 | PIK3CD | 9563 | 1550 | 1.916 | 0.5766 | Yes | ||
| 25 | TYK2 | 12058 19215 | 1683 | 1.742 | 0.5809 | Yes | ||
| 26 | SOCS3 | 20131 | 1779 | 1.638 | 0.5866 | Yes | ||
| 27 | STAM | 2912 15117 | 1995 | 1.404 | 0.5842 | No | ||
| 28 | IL10RB | 9169 | 2176 | 1.242 | 0.5827 | No | ||
| 29 | PIAS4 | 19681 | 2293 | 1.123 | 0.5838 | No | ||
| 30 | IL10 | 14145 1510 1553 22902 | 2470 | 0.996 | 0.5809 | No | ||
| 31 | IL6ST | 4920 | 2717 | 0.771 | 0.5727 | No | ||
| 32 | MYC | 22465 9435 | 3045 | 0.531 | 0.5585 | No | ||
| 33 | CCND2 | 16987 | 3097 | 0.503 | 0.5590 | No | ||
| 34 | CRLF2 | 16442 | 3339 | 0.384 | 0.5485 | No | ||
| 35 | IFNAR2 | 22705 1699 | 3380 | 0.366 | 0.5488 | No | ||
| 36 | IL7 | 4921 | 3396 | 0.362 | 0.5504 | No | ||
| 37 | PIAS3 | 15491 672 1906 | 3768 | 0.238 | 0.5319 | No | ||
| 38 | IL9R | 20505 | 3977 | 0.186 | 0.5218 | No | ||
| 39 | LEPR | 2531 2349 | 4003 | 0.181 | 0.5217 | No | ||
| 40 | IL13RA1 | 24361 | 4158 | 0.153 | 0.5143 | No | ||
| 41 | CTF1 | 18064 | 4298 | 0.135 | 0.5077 | No | ||
| 42 | BCL2L1 | 4440 2930 8652 | 4316 | 0.132 | 0.5077 | No | ||
| 43 | CLCF1 | 12160 3742 | 4347 | 0.127 | 0.5069 | No | ||
| 44 | SPRY1 | 15606 1901 | 4349 | 0.127 | 0.5077 | No | ||
| 45 | EPOR | 19204 | 4365 | 0.123 | 0.5077 | No | ||
| 46 | IFNAR1 | 22703 | 4428 | 0.116 | 0.5051 | No | ||
| 47 | PIK3R5 | 11507 | 4525 | 0.104 | 0.5006 | No | ||
| 48 | CSF2 | 20454 | 4545 | 0.101 | 0.5002 | No | ||
| 49 | CSF3R | 8804 8803 2326 4565 | 4551 | 0.100 | 0.5006 | No | ||
| 50 | IL11 | 17981 | 4577 | 0.097 | 0.4999 | No | ||
| 51 | IL4R | 18085 | 4633 | 0.093 | 0.4975 | No | ||
| 52 | SOS1 | 5476 | 4708 | 0.086 | 0.4941 | No | ||
| 53 | CSF3 | 1394 20671 | 4912 | 0.073 | 0.4836 | No | ||
| 54 | PIK3R1 | 3170 | 4927 | 0.072 | 0.4833 | No | ||
| 55 | PIAS2 | 1948 5102 | 4977 | 0.069 | 0.4811 | No | ||
| 56 | EPO | 8911 | 5245 | 0.056 | 0.4670 | No | ||
| 57 | IL20 | 13840 | 5637 | 0.042 | 0.4462 | No | ||
| 58 | IFNB1 | 15846 | 5642 | 0.042 | 0.4462 | No | ||
| 59 | IL23A | 19598 | 5852 | 0.036 | 0.4351 | No | ||
| 60 | IL23R | 17127 | 5865 | 0.036 | 0.4347 | No | ||
| 61 | IL22RA1 | 16037 | 5958 | 0.034 | 0.4300 | No | ||
| 62 | SOCS5 | 1619 23141 | 6137 | 0.030 | 0.4205 | No | ||
| 63 | TSLP | 23604 1955 | 6168 | 0.029 | 0.4191 | No | ||
| 64 | GHR | 22336 | 6278 | 0.027 | 0.4134 | No | ||
| 65 | IL22 | 11948 | 6297 | 0.026 | 0.4126 | No | ||
| 66 | IFNA13 | 15844 | 6404 | 0.024 | 0.4070 | No | ||
| 67 | SPRED2 | 8539 4332 | 6750 | 0.018 | 0.3885 | No | ||
| 68 | STAT6 | 19854 9909 | 6760 | 0.018 | 0.3881 | No | ||
| 69 | IL13RA2 | 24036 | 7113 | 0.012 | 0.3691 | No | ||
| 70 | LEP | 17505 | 7223 | 0.011 | 0.3633 | No | ||
| 71 | TPO | 21092 | 7248 | 0.010 | 0.3621 | No | ||
| 72 | IL12B | 20918 | 7261 | 0.010 | 0.3615 | No | ||
| 73 | SPRY2 | 21725 | 7274 | 0.010 | 0.3609 | No | ||
| 74 | IL21 | 12241 | 7600 | 0.005 | 0.3434 | No | ||
| 75 | PIK3CA | 9562 | 7760 | 0.003 | 0.3348 | No | ||
| 76 | PRL | 21690 | 8689 | -0.009 | 0.2846 | No | ||
| 77 | STAT5B | 20222 | 8729 | -0.010 | 0.2826 | No | ||
| 78 | IFNK | 11837 | 8932 | -0.013 | 0.2717 | No | ||
| 79 | CBLC | 17938 | 9067 | -0.015 | 0.2646 | No | ||
| 80 | IL19 | 13839 | 9097 | -0.015 | 0.2631 | No | ||
| 81 | MPL | 15780 | 9154 | -0.016 | 0.2602 | No | ||
| 82 | IFNA4 | 9150 | 9179 | -0.016 | 0.2590 | No | ||
| 83 | IL4 | 9174 | 9385 | -0.019 | 0.2480 | No | ||
| 84 | IL28RA | 10823 16038 | 9507 | -0.021 | 0.2416 | No | ||
| 85 | LIFR | 22515 9277 | 9700 | -0.024 | 0.2314 | No | ||
| 86 | PTPN11 | 5326 16391 9660 | 9715 | -0.024 | 0.2308 | No | ||
| 87 | AKT2 | 4365 4366 | 10089 | -0.030 | 0.2108 | No | ||
| 88 | IFNA7 | 9152 | 10191 | -0.032 | 0.2056 | No | ||
| 89 | IL2 | 15354 | 10229 | -0.032 | 0.2038 | No | ||
| 90 | IL9 | 21444 | 10317 | -0.034 | 0.1993 | No | ||
| 91 | IL5RA | 17051 | 10518 | -0.037 | 0.1887 | No | ||
| 92 | SPRY4 | 23448 | 10528 | -0.037 | 0.1885 | No | ||
| 93 | IFNA5 | 9151 | 10651 | -0.039 | 0.1821 | No | ||
| 94 | IL6 | 16895 | 10906 | -0.045 | 0.1687 | No | ||
| 95 | IL3 | 20453 | 11256 | -0.052 | 0.1502 | No | ||
| 96 | IL15RA | 2899 2717 15118 2792 | 11460 | -0.056 | 0.1395 | No | ||
| 97 | IFNA2 | 9149 | 11540 | -0.058 | 0.1357 | No | ||
| 98 | CNTF | 3741 8761 3680 3736 | 11760 | -0.064 | 0.1242 | No | ||
| 99 | JAK2 | 23893 9197 3706 | 11817 | -0.066 | 0.1216 | No | ||
| 100 | IFNG | 19869 | 12487 | -0.093 | 0.0861 | No | ||
| 101 | OSMR | 22333 | 12578 | -0.097 | 0.0818 | No | ||
| 102 | IFNA14 | 11917 | 12583 | -0.097 | 0.0822 | No | ||
| 103 | SOCS7 | 5308 20679 | 13116 | -0.126 | 0.0543 | No | ||
| 104 | AKT1 | 8568 | 13190 | -0.131 | 0.0512 | No | ||
| 105 | IL5 | 20884 10220 | 13213 | -0.133 | 0.0509 | No | ||
| 106 | IL22RA2 | 20082 | 13229 | -0.134 | 0.0510 | No | ||
| 107 | STAM2 | 7095 2845 | 13282 | -0.138 | 0.0491 | No | ||
| 108 | CNTFR | 2515 15906 | 13292 | -0.138 | 0.0495 | No | ||
| 109 | IL24 | 976 13841 | 13475 | -0.153 | 0.0407 | No | ||
| 110 | IL15 | 18826 3801 | 13490 | -0.155 | 0.0409 | No | ||
| 111 | IL6R | 1862 4919 | 13589 | -0.164 | 0.0367 | No | ||
| 112 | IFNA6 | 17749 | 13620 | -0.167 | 0.0362 | No | ||
| 113 | IL12RB2 | 17128 | 13645 | -0.169 | 0.0360 | No | ||
| 114 | IFNA1 | 9147 | 13656 | -0.170 | 0.0366 | No | ||
| 115 | CBL | 19154 | 13840 | -0.189 | 0.0279 | No | ||
| 116 | IFNE1 | 15842 | 14294 | -0.246 | 0.0050 | No | ||
| 117 | LIF | 956 667 20960 | 14556 | -0.293 | -0.0071 | No | ||
| 118 | PRLR | 9620 5291 | 14991 | -0.411 | -0.0279 | No | ||
| 119 | IL2RG | 24096 | 15069 | -0.442 | -0.0292 | No | ||
| 120 | IL21R | 1398 18084 | 15273 | -0.523 | -0.0367 | No | ||
| 121 | SPRED1 | 4331 | 15415 | -0.601 | -0.0404 | No | ||
| 122 | IL13 | 20461 | 15446 | -0.627 | -0.0378 | No | ||
| 123 | IL2RB | 22219 | 15966 | -0.989 | -0.0594 | No | ||
| 124 | PIK3R2 | 18850 | 16083 | -1.041 | -0.0588 | No | ||
| 125 | IL12RB1 | 3916 18582 | 16155 | -1.092 | -0.0554 | No | ||
| 126 | SOCS4 | 7424 | 16918 | -1.910 | -0.0841 | No | ||
| 127 | SOCS1 | 4522 | 17035 | -2.077 | -0.0766 | No | ||
| 128 | IL10RA | 19137 | 17083 | -2.152 | -0.0650 | No | ||
| 129 | IL2RA | 4918 | 17354 | -2.627 | -0.0623 | No | ||
| 130 | PIK3R3 | 5248 | 17678 | -3.275 | -0.0581 | No | ||
| 131 | PTPN6 | 17002 | 17810 | -3.582 | -0.0416 | No | ||
| 132 | CCND1 | 4487 4488 8707 17535 | 17818 | -3.619 | -0.0181 | No | ||
| 133 | PIK3CB | 19030 | 18080 | -4.515 | -0.0024 | No | ||
| 134 | CBLB | 5531 22734 | 18142 | -4.749 | 0.0256 | No |