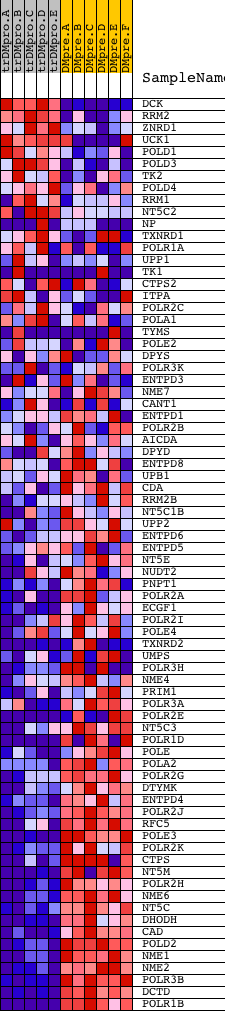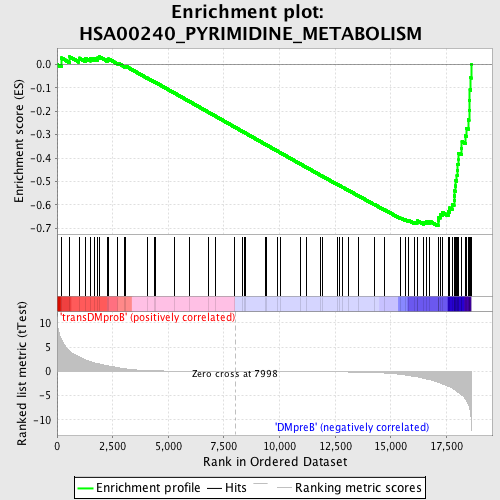
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | HSA00240_PYRIMIDINE_METABOLISM |
| Enrichment Score (ES) | -0.6902056 |
| Normalized Enrichment Score (NES) | -1.6664953 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0313999 |
| FWER p-Value | 0.13 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DCK | 16808 | 192 | 6.569 | 0.0282 | No | ||
| 2 | RRM2 | 5401 5400 | 571 | 4.235 | 0.0326 | No | ||
| 3 | ZNRD1 | 1491 22987 | 983 | 2.983 | 0.0280 | No | ||
| 4 | UCK1 | 10254 5831 10253 | 1283 | 2.344 | 0.0256 | No | ||
| 5 | POLD1 | 17847 | 1481 | 2.020 | 0.0268 | No | ||
| 6 | POLD3 | 17742 | 1682 | 1.745 | 0.0263 | No | ||
| 7 | TK2 | 18779 | 1824 | 1.608 | 0.0281 | No | ||
| 8 | POLD4 | 12822 | 1902 | 1.515 | 0.0328 | No | ||
| 9 | RRM1 | 18163 | 2252 | 1.164 | 0.0208 | No | ||
| 10 | NT5C2 | 3768 8052 | 2316 | 1.102 | 0.0239 | No | ||
| 11 | NP | 22027 9597 5273 5274 | 2722 | 0.766 | 0.0065 | No | ||
| 12 | TXNRD1 | 19923 | 3048 | 0.530 | -0.0079 | No | ||
| 13 | POLR1A | 9749 5393 | 3076 | 0.513 | -0.0063 | No | ||
| 14 | UPP1 | 20947 1385 | 4044 | 0.173 | -0.0575 | No | ||
| 15 | TK1 | 1457 10182 5762 | 4377 | 0.122 | -0.0747 | No | ||
| 16 | CTPS2 | 12060 2571 | 4426 | 0.116 | -0.0766 | No | ||
| 17 | ITPA | 9193 | 5267 | 0.056 | -0.1215 | No | ||
| 18 | POLR2C | 9750 | 5936 | 0.034 | -0.1574 | No | ||
| 19 | POLA1 | 24112 | 6819 | 0.017 | -0.2048 | No | ||
| 20 | TYMS | 5810 5809 3606 3598 | 7118 | 0.012 | -0.2208 | No | ||
| 21 | POLE2 | 21053 | 7971 | 0.000 | -0.2668 | No | ||
| 22 | DPYS | 22305 | 8327 | -0.005 | -0.2859 | No | ||
| 23 | POLR3K | 12447 7372 | 8411 | -0.006 | -0.2904 | No | ||
| 24 | ENTPD3 | 19266 | 8468 | -0.006 | -0.2933 | No | ||
| 25 | NME7 | 926 4101 14070 3986 3946 931 | 9369 | -0.019 | -0.3418 | No | ||
| 26 | CANT1 | 13304 | 9398 | -0.019 | -0.3432 | No | ||
| 27 | ENTPD1 | 4495 | 9917 | -0.027 | -0.3709 | No | ||
| 28 | POLR2B | 16817 | 10054 | -0.030 | -0.3781 | No | ||
| 29 | AICDA | 17295 1087 | 10920 | -0.045 | -0.4245 | No | ||
| 30 | DPYD | 15437 | 11226 | -0.051 | -0.4406 | No | ||
| 31 | ENTPD8 | 12996 | 11840 | -0.067 | -0.4733 | No | ||
| 32 | UPB1 | 19988 3389 | 11908 | -0.069 | -0.4765 | No | ||
| 33 | CDA | 15702 | 12600 | -0.097 | -0.5132 | No | ||
| 34 | RRM2B | 6902 6903 11801 | 12691 | -0.101 | -0.5175 | No | ||
| 35 | NT5C1B | 21315 | 12830 | -0.109 | -0.5243 | No | ||
| 36 | UPP2 | 15009 13346 8032 | 13109 | -0.125 | -0.5385 | No | ||
| 37 | ENTPD6 | 4496 2678 | 13526 | -0.158 | -0.5601 | No | ||
| 38 | ENTPD5 | 4497 | 14246 | -0.240 | -0.5974 | No | ||
| 39 | NT5E | 19360 18702 | 14728 | -0.340 | -0.6214 | No | ||
| 40 | NUDT2 | 16239 | 15441 | -0.622 | -0.6561 | No | ||
| 41 | PNPT1 | 20936 | 15655 | -0.746 | -0.6632 | No | ||
| 42 | POLR2A | 5394 | 15814 | -0.870 | -0.6666 | No | ||
| 43 | ECGF1 | 22160 | 16080 | -1.039 | -0.6748 | No | ||
| 44 | POLR2I | 12839 | 16178 | -1.117 | -0.6735 | No | ||
| 45 | POLE4 | 12441 | 16204 | -1.143 | -0.6682 | No | ||
| 46 | TXNRD2 | 22829 1649 | 16488 | -1.445 | -0.6750 | No | ||
| 47 | UMPS | 22606 1760 | 16613 | -1.563 | -0.6725 | No | ||
| 48 | POLR3H | 13460 8135 | 16747 | -1.684 | -0.6698 | No | ||
| 49 | NME4 | 23065 1577 | 17127 | -2.204 | -0.6773 | Yes | ||
| 50 | PRIM1 | 19847 | 17128 | -2.204 | -0.6643 | Yes | ||
| 51 | POLR3A | 21900 | 17155 | -2.229 | -0.6527 | Yes | ||
| 52 | POLR2E | 3325 19699 | 17221 | -2.334 | -0.6425 | Yes | ||
| 53 | NT5C3 | 17136 | 17316 | -2.573 | -0.6325 | Yes | ||
| 54 | POLR1D | 3593 3658 16623 | 17578 | -3.037 | -0.6287 | Yes | ||
| 55 | POLE | 16755 | 17631 | -3.130 | -0.6132 | Yes | ||
| 56 | POLA2 | 23988 | 17757 | -3.452 | -0.5997 | Yes | ||
| 57 | POLR2G | 23753 | 17842 | -3.679 | -0.5826 | Yes | ||
| 58 | DTYMK | 5776 | 17856 | -3.721 | -0.5615 | Yes | ||
| 59 | ENTPD4 | 7449 | 17875 | -3.773 | -0.5403 | Yes | ||
| 60 | POLR2J | 16672 | 17905 | -3.892 | -0.5191 | Yes | ||
| 61 | RFC5 | 13005 7791 | 17920 | -3.951 | -0.4966 | Yes | ||
| 62 | POLE3 | 7200 | 17972 | -4.110 | -0.4753 | Yes | ||
| 63 | POLR2K | 9413 | 17988 | -4.160 | -0.4517 | Yes | ||
| 64 | CTPS | 2514 15772 | 18007 | -4.223 | -0.4279 | Yes | ||
| 65 | NT5M | 8345 4175 | 18037 | -4.318 | -0.4041 | Yes | ||
| 66 | POLR2H | 10888 | 18043 | -4.355 | -0.3789 | Yes | ||
| 67 | NME6 | 3139 19297 | 18170 | -4.853 | -0.3572 | Yes | ||
| 68 | NT5C | 20151 | 18198 | -4.950 | -0.3296 | Yes | ||
| 69 | DHODH | 7152 | 18357 | -5.661 | -0.3049 | Yes | ||
| 70 | CAD | 16886 | 18408 | -6.035 | -0.2722 | Yes | ||
| 71 | POLD2 | 20537 | 18484 | -6.826 | -0.2362 | Yes | ||
| 72 | NME1 | 9467 | 18515 | -7.106 | -0.1962 | Yes | ||
| 73 | NME2 | 9468 | 18538 | -7.449 | -0.1537 | Yes | ||
| 74 | POLR3B | 12875 | 18557 | -7.903 | -0.1083 | Yes | ||
| 75 | DCTD | 18620 | 18594 | -9.062 | -0.0571 | Yes | ||
| 76 | POLR1B | 14857 | 18608 | -9.919 | 0.0004 | Yes |