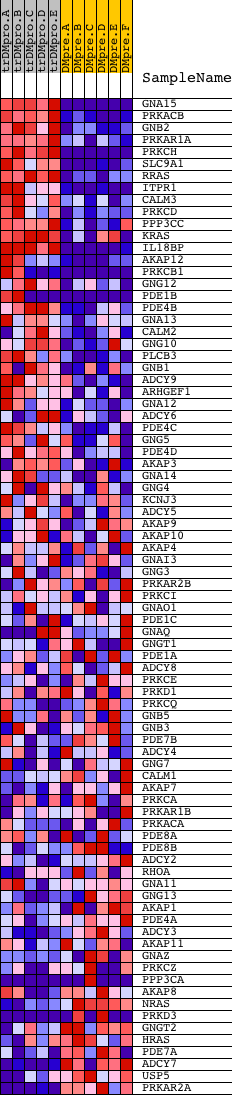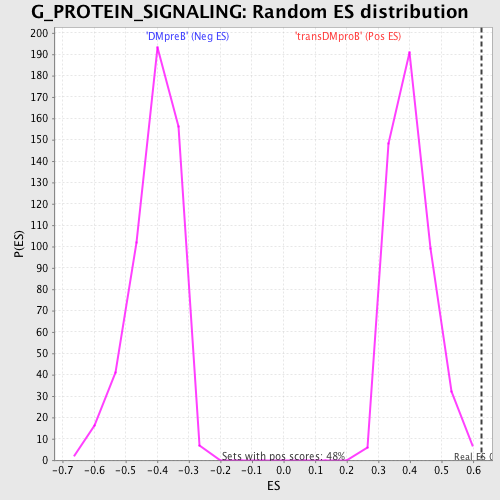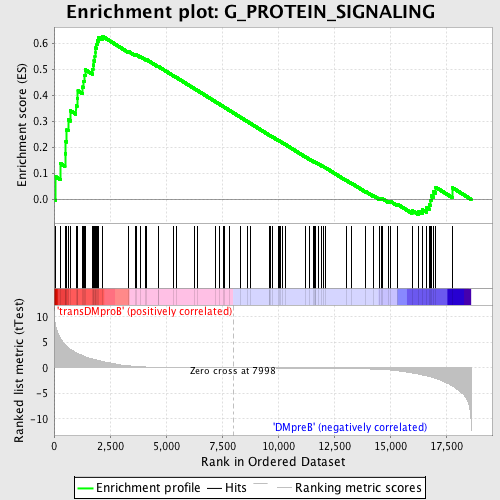
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | G_PROTEIN_SIGNALING |
| Enrichment Score (ES) | 0.6275508 |
| Normalized Enrichment Score (NES) | 1.5602696 |
| Nominal p-value | 0.0020703934 |
| FDR q-value | 0.14926912 |
| FWER p-Value | 0.803 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GNA15 | 19671 | 61 | 8.343 | 0.0876 | Yes | ||
| 2 | PRKACB | 15140 | 286 | 5.707 | 0.1378 | Yes | ||
| 3 | GNB2 | 9026 | 487 | 4.520 | 0.1762 | Yes | ||
| 4 | PRKAR1A | 20615 | 522 | 4.403 | 0.2224 | Yes | ||
| 5 | PRKCH | 21246 | 560 | 4.274 | 0.2670 | Yes | ||
| 6 | SLC9A1 | 16053 | 628 | 3.989 | 0.3068 | Yes | ||
| 7 | RRAS | 18256 | 735 | 3.612 | 0.3405 | Yes | ||
| 8 | ITPR1 | 17341 | 977 | 2.997 | 0.3601 | Yes | ||
| 9 | CALM3 | 8682 | 1025 | 2.875 | 0.3890 | Yes | ||
| 10 | PRKCD | 21897 | 1065 | 2.795 | 0.4173 | Yes | ||
| 11 | PPP3CC | 21763 | 1270 | 2.374 | 0.4322 | Yes | ||
| 12 | KRAS | 9247 | 1320 | 2.284 | 0.4544 | Yes | ||
| 13 | IL18BP | 17730 | 1363 | 2.208 | 0.4762 | Yes | ||
| 14 | AKAP12 | 17600 | 1392 | 2.167 | 0.4984 | Yes | ||
| 15 | PRKCB1 | 1693 9574 | 1709 | 1.712 | 0.5000 | Yes | ||
| 16 | GNG12 | 4789 | 1739 | 1.677 | 0.5167 | Yes | ||
| 17 | PDE1B | 9542 | 1778 | 1.639 | 0.5325 | Yes | ||
| 18 | PDE4B | 9543 | 1822 | 1.611 | 0.5477 | Yes | ||
| 19 | GNA13 | 20617 | 1827 | 1.605 | 0.5650 | Yes | ||
| 20 | CALM2 | 8681 | 1832 | 1.596 | 0.5822 | Yes | ||
| 21 | GNG10 | 4788 | 1904 | 1.514 | 0.5949 | Yes | ||
| 22 | PLCB3 | 23799 | 1926 | 1.484 | 0.6099 | Yes | ||
| 23 | GNB1 | 15967 | 1980 | 1.431 | 0.6226 | Yes | ||
| 24 | ADCY9 | 22680 | 2147 | 1.272 | 0.6276 | Yes | ||
| 25 | ARHGEF1 | 1952 18343 3996 | 3312 | 0.395 | 0.5691 | No | ||
| 26 | GNA12 | 9023 | 3615 | 0.280 | 0.5558 | No | ||
| 27 | ADCY6 | 22139 2283 8551 | 3656 | 0.270 | 0.5566 | No | ||
| 28 | PDE4C | 8481 | 3857 | 0.217 | 0.5482 | No | ||
| 29 | GNG5 | 9030 15393 483 | 4091 | 0.165 | 0.5374 | No | ||
| 30 | PDE4D | 10722 6235 | 4135 | 0.158 | 0.5368 | No | ||
| 31 | AKAP3 | 8566 | 4646 | 0.092 | 0.5103 | No | ||
| 32 | GNA14 | 23907 | 4649 | 0.091 | 0.5112 | No | ||
| 33 | GNG4 | 21706 | 5348 | 0.052 | 0.4741 | No | ||
| 34 | KCNJ3 | 4944 | 5474 | 0.047 | 0.4678 | No | ||
| 35 | ADCY5 | 10312 | 6281 | 0.027 | 0.4246 | No | ||
| 36 | AKAP9 | 16604 16603 4150 8303 | 6422 | 0.024 | 0.4173 | No | ||
| 37 | AKAP10 | 20411 7145 | 7190 | 0.011 | 0.3761 | No | ||
| 38 | AKAP4 | 24394 2602 | 7363 | 0.008 | 0.3669 | No | ||
| 39 | GNAI3 | 15198 | 7576 | 0.005 | 0.3555 | No | ||
| 40 | GNG3 | 23752 | 7624 | 0.005 | 0.3530 | No | ||
| 41 | PRKAR2B | 5288 2107 | 7813 | 0.002 | 0.3429 | No | ||
| 42 | PRKCI | 9576 | 8313 | -0.004 | 0.3160 | No | ||
| 43 | GNAO1 | 4785 3829 | 8644 | -0.009 | 0.2983 | No | ||
| 44 | PDE1C | 5235 | 8764 | -0.010 | 0.2920 | No | ||
| 45 | GNAQ | 4786 23909 3685 | 9625 | -0.023 | 0.2459 | No | ||
| 46 | GNGT1 | 17533 | 9636 | -0.023 | 0.2456 | No | ||
| 47 | PDE1A | 9541 5234 | 9653 | -0.023 | 0.2450 | No | ||
| 48 | ADCY8 | 22281 | 9741 | -0.024 | 0.2405 | No | ||
| 49 | PRKCE | 9575 | 10004 | -0.029 | 0.2267 | No | ||
| 50 | PRKD1 | 2074 21075 | 10042 | -0.029 | 0.2250 | No | ||
| 51 | PRKCQ | 2873 2831 | 10099 | -0.030 | 0.2223 | No | ||
| 52 | GNB5 | 2985 9029 3153 | 10186 | -0.032 | 0.2180 | No | ||
| 53 | GNB3 | 9027 | 10337 | -0.034 | 0.2103 | No | ||
| 54 | PDE7B | 19807 | 11236 | -0.051 | 0.1624 | No | ||
| 55 | ADCY4 | 2980 21818 | 11398 | -0.055 | 0.1543 | No | ||
| 56 | GNG7 | 19684 | 11595 | -0.060 | 0.1444 | No | ||
| 57 | CALM1 | 21184 | 11604 | -0.060 | 0.1446 | No | ||
| 58 | AKAP7 | 6478 | 11663 | -0.062 | 0.1422 | No | ||
| 59 | PRKCA | 20174 | 11681 | -0.062 | 0.1419 | No | ||
| 60 | PRKAR1B | 16320 | 11809 | -0.066 | 0.1358 | No | ||
| 61 | PRKACA | 18549 3844 | 11953 | -0.071 | 0.1289 | No | ||
| 62 | PDE8A | 18202 | 12014 | -0.073 | 0.1264 | No | ||
| 63 | PDE8B | 5756 | 12107 | -0.076 | 0.1223 | No | ||
| 64 | ADCY2 | 21412 | 13035 | -0.121 | 0.0736 | No | ||
| 65 | RHOA | 8624 4409 4410 | 13287 | -0.138 | 0.0616 | No | ||
| 66 | GNA11 | 3300 3345 19670 | 13911 | -0.196 | 0.0301 | No | ||
| 67 | GNG13 | 23338 | 14257 | -0.241 | 0.0141 | No | ||
| 68 | AKAP1 | 4364 1326 1286 20299 | 14520 | -0.287 | 0.0031 | No | ||
| 69 | PDE4A | 3037 19542 3083 3023 | 14605 | -0.307 | 0.0019 | No | ||
| 70 | ADCY3 | 21329 894 | 14643 | -0.316 | 0.0033 | No | ||
| 71 | AKAP11 | 2695 | 14944 | -0.400 | -0.0085 | No | ||
| 72 | GNAZ | 71 | 15030 | -0.424 | -0.0084 | No | ||
| 73 | PRKCZ | 5260 | 15328 | -0.546 | -0.0185 | No | ||
| 74 | PPP3CA | 1863 5284 | 15979 | -0.995 | -0.0427 | No | ||
| 75 | AKAP8 | 12121 | 16260 | -1.193 | -0.0448 | No | ||
| 76 | NRAS | 5191 | 16449 | -1.395 | -0.0398 | No | ||
| 77 | PRKD3 | 13252 7956 | 16622 | -1.570 | -0.0319 | No | ||
| 78 | GNGT2 | 20691 | 16745 | -1.681 | -0.0202 | No | ||
| 79 | HRAS | 4868 | 16802 | -1.753 | -0.0041 | No | ||
| 80 | PDE7A | 9544 1788 | 16836 | -1.794 | 0.0136 | No | ||
| 81 | ADCY7 | 8552 448 | 16937 | -1.943 | 0.0294 | No | ||
| 82 | USP5 | 17004 | 17036 | -2.078 | 0.0468 | No | ||
| 83 | PRKAR2A | 5287 | 17789 | -3.523 | 0.0446 | No |