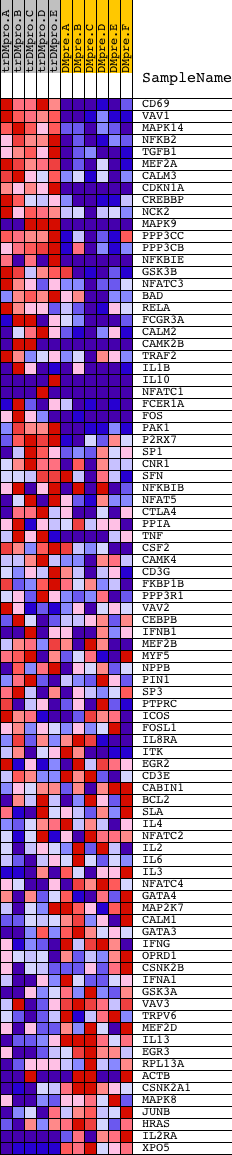
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
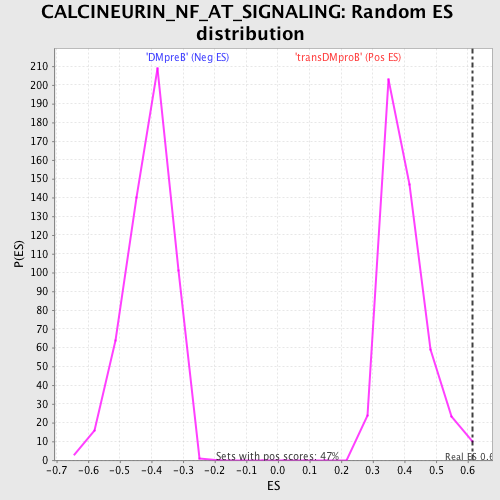
| GeneSet | CALCINEURIN_NF_AT_SIGNALING |
| Enrichment Score (ES) | 0.61555624 |
| Normalized Enrichment Score (NES) | 1.5438639 |
| Nominal p-value | 0.006437768 |
| FDR q-value | 0.16007176 |
| FWER p-Value | 0.876 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CD69 | 16979 8717 | 185 | 6.606 | 0.0642 | Yes | ||
| 2 | VAV1 | 23173 | 347 | 5.300 | 0.1150 | Yes | ||
| 3 | MAPK14 | 23313 | 429 | 4.797 | 0.1644 | Yes | ||
| 4 | NFKB2 | 23810 | 491 | 4.513 | 0.2118 | Yes | ||
| 5 | TGFB1 | 18332 | 616 | 4.024 | 0.2502 | Yes | ||
| 6 | MEF2A | 1201 5089 3492 | 854 | 3.297 | 0.2745 | Yes | ||
| 7 | CALM3 | 8682 | 1025 | 2.875 | 0.2976 | Yes | ||
| 8 | CDKN1A | 4511 8729 | 1034 | 2.865 | 0.3293 | Yes | ||
| 9 | CREBBP | 22682 8783 | 1098 | 2.720 | 0.3564 | Yes | ||
| 10 | NCK2 | 9448 | 1208 | 2.518 | 0.3788 | Yes | ||
| 11 | MAPK9 | 1233 20903 1383 | 1236 | 2.449 | 0.4048 | Yes | ||
| 12 | PPP3CC | 21763 | 1270 | 2.374 | 0.4297 | Yes | ||
| 13 | PPP3CB | 5285 | 1370 | 2.196 | 0.4490 | Yes | ||
| 14 | NFKBIE | 23225 1556 | 1402 | 2.139 | 0.4713 | Yes | ||
| 15 | GSK3B | 22761 | 1471 | 2.035 | 0.4905 | Yes | ||
| 16 | NFATC3 | 5169 | 1511 | 1.983 | 0.5106 | Yes | ||
| 17 | BAD | 24000 | 1588 | 1.876 | 0.5276 | Yes | ||
| 18 | RELA | 23783 | 1612 | 1.845 | 0.5470 | Yes | ||
| 19 | FCGR3A | 8960 | 1655 | 1.782 | 0.5648 | Yes | ||
| 20 | CALM2 | 8681 | 1832 | 1.596 | 0.5732 | Yes | ||
| 21 | CAMK2B | 20536 | 1834 | 1.586 | 0.5909 | Yes | ||
| 22 | TRAF2 | 14657 | 2284 | 1.132 | 0.5794 | Yes | ||
| 23 | IL1B | 14431 | 2370 | 1.061 | 0.5867 | Yes | ||
| 24 | IL10 | 14145 1510 1553 22902 | 2470 | 0.996 | 0.5926 | Yes | ||
| 25 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 2476 | 0.992 | 0.6034 | Yes | ||
| 26 | FCER1A | 8957 | 2492 | 0.979 | 0.6136 | Yes | ||
| 27 | FOS | 21202 | 2723 | 0.766 | 0.6098 | Yes | ||
| 28 | PAK1 | 9527 | 2767 | 0.720 | 0.6156 | Yes | ||
| 29 | P2RX7 | 3487 9520 487 | 3248 | 0.425 | 0.5944 | No | ||
| 30 | SP1 | 9852 | 3303 | 0.399 | 0.5960 | No | ||
| 31 | CNR1 | 4537 2362 | 3574 | 0.291 | 0.5847 | No | ||
| 32 | SFN | 7060 12062 | 3710 | 0.254 | 0.5803 | No | ||
| 33 | NFKBIB | 17906 | 4009 | 0.178 | 0.5662 | No | ||
| 34 | NFAT5 | 3921 7037 12036 | 4055 | 0.171 | 0.5657 | No | ||
| 35 | CTLA4 | 14238 | 4056 | 0.171 | 0.5676 | No | ||
| 36 | PPIA | 1284 11188 | 4376 | 0.122 | 0.5517 | No | ||
| 37 | TNF | 23004 | 4390 | 0.120 | 0.5524 | No | ||
| 38 | CSF2 | 20454 | 4545 | 0.101 | 0.5452 | No | ||
| 39 | CAMK4 | 4473 | 4707 | 0.086 | 0.5375 | No | ||
| 40 | CD3G | 19139 | 4851 | 0.076 | 0.5306 | No | ||
| 41 | FKBP1B | 21116 | 5071 | 0.065 | 0.5195 | No | ||
| 42 | PPP3R1 | 9611 489 | 5287 | 0.055 | 0.5085 | No | ||
| 43 | VAV2 | 5848 2670 | 5446 | 0.048 | 0.5005 | No | ||
| 44 | CEBPB | 8733 | 5620 | 0.042 | 0.4917 | No | ||
| 45 | IFNB1 | 15846 | 5642 | 0.042 | 0.4910 | No | ||
| 46 | MEF2B | 9377 | 5657 | 0.042 | 0.4907 | No | ||
| 47 | MYF5 | 19632 | 6001 | 0.033 | 0.4726 | No | ||
| 48 | NPPB | 15994 | 6427 | 0.024 | 0.4499 | No | ||
| 49 | PIN1 | 19548 | 6838 | 0.016 | 0.4280 | No | ||
| 50 | SP3 | 5483 | 6951 | 0.015 | 0.4221 | No | ||
| 51 | PTPRC | 5327 9662 | 6953 | 0.015 | 0.4222 | No | ||
| 52 | ICOS | 12013 | 7389 | 0.008 | 0.3988 | No | ||
| 53 | FOSL1 | 23779 | 7606 | 0.005 | 0.3872 | No | ||
| 54 | IL8RA | 13925 | 8052 | -0.001 | 0.3632 | No | ||
| 55 | ITK | 4934 | 8570 | -0.008 | 0.3354 | No | ||
| 56 | EGR2 | 8886 | 8783 | -0.011 | 0.3241 | No | ||
| 57 | CD3E | 8714 | 8819 | -0.011 | 0.3223 | No | ||
| 58 | CABIN1 | 19733 | 8892 | -0.012 | 0.3185 | No | ||
| 59 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 9115 | -0.015 | 0.3067 | No | ||
| 60 | SLA | 5449 | 9234 | -0.017 | 0.3006 | No | ||
| 61 | IL4 | 9174 | 9385 | -0.019 | 0.2927 | No | ||
| 62 | NFATC2 | 5168 2866 | 9389 | -0.019 | 0.2927 | No | ||
| 63 | IL2 | 15354 | 10229 | -0.032 | 0.2478 | No | ||
| 64 | IL6 | 16895 | 10906 | -0.045 | 0.2118 | No | ||
| 65 | IL3 | 20453 | 11256 | -0.052 | 0.1936 | No | ||
| 66 | NFATC4 | 22002 | 11337 | -0.054 | 0.1899 | No | ||
| 67 | GATA4 | 21792 4755 | 11466 | -0.056 | 0.1836 | No | ||
| 68 | MAP2K7 | 6453 | 11507 | -0.057 | 0.1821 | No | ||
| 69 | CALM1 | 21184 | 11604 | -0.060 | 0.1776 | No | ||
| 70 | GATA3 | 9004 | 11758 | -0.064 | 0.1700 | No | ||
| 71 | IFNG | 19869 | 12487 | -0.093 | 0.1318 | No | ||
| 72 | OPRD1 | 15737 | 13108 | -0.125 | 0.0997 | No | ||
| 73 | CSNK2B | 23008 | 13539 | -0.159 | 0.0783 | No | ||
| 74 | IFNA1 | 9147 | 13656 | -0.170 | 0.0740 | No | ||
| 75 | GSK3A | 405 | 14455 | -0.274 | 0.0340 | No | ||
| 76 | VAV3 | 1774 1848 15443 | 14752 | -0.346 | 0.0219 | No | ||
| 77 | TRPV6 | 17171 | 14770 | -0.350 | 0.0249 | No | ||
| 78 | MEF2D | 9379 | 14907 | -0.385 | 0.0219 | No | ||
| 79 | IL13 | 20461 | 15446 | -0.627 | -0.0001 | No | ||
| 80 | EGR3 | 4656 | 15650 | -0.741 | -0.0028 | No | ||
| 81 | RPL13A | 10226 79 | 15871 | -0.912 | -0.0044 | No | ||
| 82 | ACTB | 8534 337 337 338 | 15929 | -0.962 | 0.0033 | No | ||
| 83 | CSNK2A1 | 14797 | 15954 | -0.982 | 0.0130 | No | ||
| 84 | MAPK8 | 6459 | 16452 | -1.403 | 0.0019 | No | ||
| 85 | JUNB | 9201 | 16588 | -1.542 | 0.0120 | No | ||
| 86 | HRAS | 4868 | 16802 | -1.753 | 0.0202 | No | ||
| 87 | IL2RA | 4918 | 17354 | -2.627 | 0.0199 | No | ||
| 88 | XPO5 | 7802 | 18027 | -4.291 | 0.0318 | No |