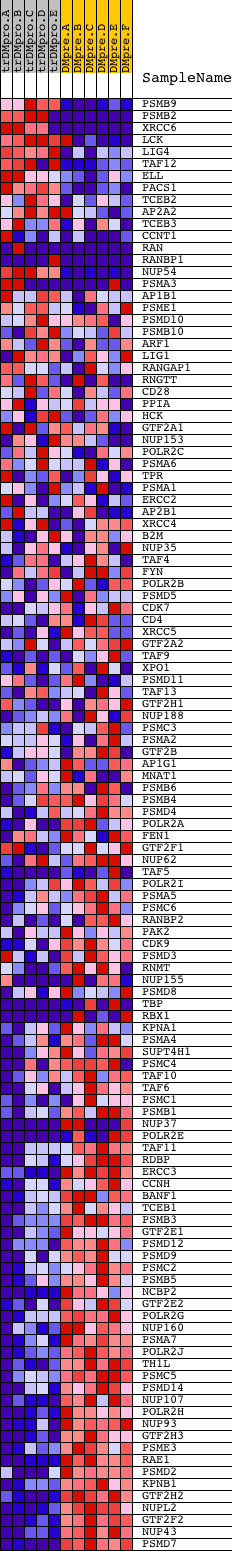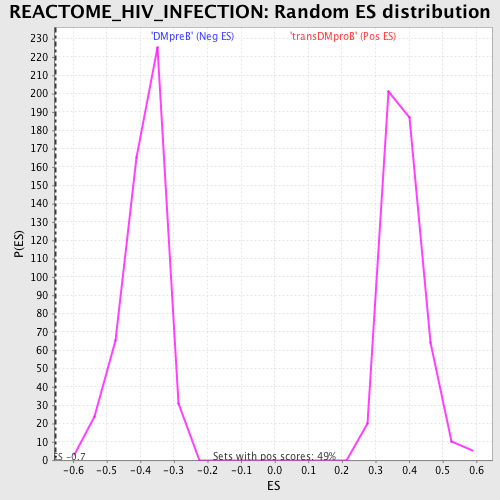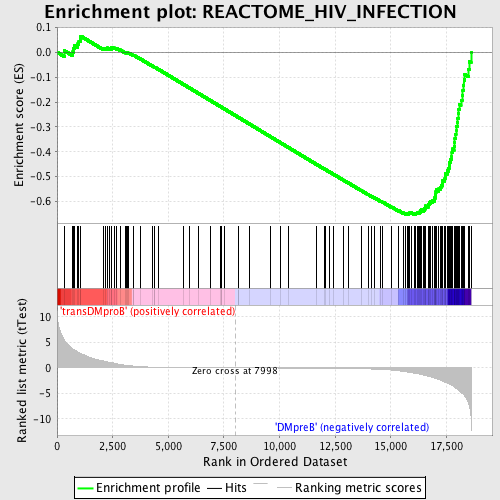
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | REACTOME_HIV_INFECTION |
| Enrichment Score (ES) | -0.65191853 |
| Normalized Enrichment Score (NES) | -1.6751153 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.018967876 |
| FWER p-Value | 0.199 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMB9 | 23021 | 323 | 5.459 | 0.0063 | No | ||
| 2 | PSMB2 | 2324 16078 | 711 | 3.670 | 0.0014 | No | ||
| 3 | XRCC6 | 2286 8991 | 742 | 3.590 | 0.0154 | No | ||
| 4 | LCK | 15746 | 786 | 3.491 | 0.0283 | No | ||
| 5 | LIG4 | 11454 | 935 | 3.083 | 0.0338 | No | ||
| 6 | TAF12 | 2537 16058 2338 | 979 | 2.991 | 0.0445 | No | ||
| 7 | ELL | 18586 | 1041 | 2.850 | 0.0536 | No | ||
| 8 | PACS1 | 23975 | 1062 | 2.803 | 0.0647 | No | ||
| 9 | TCEB2 | 12563 23102 7476 | 2076 | 1.333 | 0.0158 | No | ||
| 10 | AP2A2 | 18009 | 2196 | 1.223 | 0.0147 | No | ||
| 11 | TCEB3 | 15707 | 2243 | 1.172 | 0.0173 | No | ||
| 12 | CCNT1 | 22140 11607 | 2349 | 1.081 | 0.0163 | No | ||
| 13 | RAN | 5356 9691 | 2444 | 1.000 | 0.0156 | No | ||
| 14 | RANBP1 | 9692 5357 | 2445 | 1.000 | 0.0200 | No | ||
| 15 | NUP54 | 11231 11232 6516 | 2571 | 0.901 | 0.0171 | No | ||
| 16 | PSMA3 | 9632 5298 | 2673 | 0.808 | 0.0152 | No | ||
| 17 | AP1B1 | 8599 | 2838 | 0.661 | 0.0092 | No | ||
| 18 | PSME1 | 5300 9637 | 3070 | 0.516 | -0.0010 | No | ||
| 19 | PSMD10 | 2578 2597 24046 | 3127 | 0.485 | -0.0019 | No | ||
| 20 | PSMB10 | 5299 18761 | 3162 | 0.463 | -0.0018 | No | ||
| 21 | ARF1 | 64 | 3224 | 0.439 | -0.0032 | No | ||
| 22 | LIG1 | 18388 1749 1493 | 3446 | 0.342 | -0.0136 | No | ||
| 23 | RANGAP1 | 2180 22195 | 3447 | 0.342 | -0.0121 | No | ||
| 24 | RNGTT | 10769 2354 6272 | 3737 | 0.247 | -0.0267 | No | ||
| 25 | CD28 | 14239 4092 | 4275 | 0.138 | -0.0551 | No | ||
| 26 | PPIA | 1284 11188 | 4376 | 0.122 | -0.0600 | No | ||
| 27 | HCK | 14787 | 4391 | 0.120 | -0.0602 | No | ||
| 28 | GTF2A1 | 2064 8187 8188 13512 | 4575 | 0.098 | -0.0697 | No | ||
| 29 | NUP153 | 21474 | 5681 | 0.041 | -0.1292 | No | ||
| 30 | POLR2C | 9750 | 5936 | 0.034 | -0.1428 | No | ||
| 31 | PSMA6 | 21270 | 6335 | 0.025 | -0.1642 | No | ||
| 32 | TPR | 927 4255 | 6877 | 0.016 | -0.1934 | No | ||
| 33 | PSMA1 | 1627 17669 | 7334 | 0.009 | -0.2180 | No | ||
| 34 | ERCC2 | 1549 18363 3812 | 7384 | 0.008 | -0.2206 | No | ||
| 35 | AP2B1 | 12958 7752 12957 1280 | 7401 | 0.008 | -0.2215 | No | ||
| 36 | XRCC4 | 4238 8432 | 7532 | 0.006 | -0.2285 | No | ||
| 37 | B2M | 72 14882 | 8147 | -0.002 | -0.2617 | No | ||
| 38 | NUP35 | 12803 | 8155 | -0.002 | -0.2620 | No | ||
| 39 | TAF4 | 14319 | 8626 | -0.008 | -0.2874 | No | ||
| 40 | FYN | 3375 3395 20052 | 9588 | -0.022 | -0.3393 | No | ||
| 41 | POLR2B | 16817 | 10054 | -0.030 | -0.3643 | No | ||
| 42 | PSMD5 | 2774 14612 | 10407 | -0.035 | -0.3832 | No | ||
| 43 | CDK7 | 21365 | 11650 | -0.061 | -0.4500 | No | ||
| 44 | CD4 | 16999 | 12030 | -0.073 | -0.4702 | No | ||
| 45 | XRCC5 | 14229 | 12055 | -0.074 | -0.4712 | No | ||
| 46 | GTF2A2 | 10654 | 12265 | -0.082 | -0.4821 | No | ||
| 47 | TAF9 | 3213 8433 | 12406 | -0.089 | -0.4893 | No | ||
| 48 | XPO1 | 4172 | 12880 | -0.112 | -0.5144 | No | ||
| 49 | PSMD11 | 12772 7600 | 13078 | -0.123 | -0.5245 | No | ||
| 50 | TAF13 | 8288 | 13683 | -0.173 | -0.5564 | No | ||
| 51 | GTF2H1 | 4069 18236 | 13978 | -0.204 | -0.5714 | No | ||
| 52 | NUP188 | 15053 | 14132 | -0.223 | -0.5787 | No | ||
| 53 | PSMC3 | 9636 | 14271 | -0.243 | -0.5851 | No | ||
| 54 | PSMA2 | 9631 | 14554 | -0.293 | -0.5991 | No | ||
| 55 | GTF2B | 10489 | 14646 | -0.316 | -0.6026 | No | ||
| 56 | AP1G1 | 4392 | 15035 | -0.425 | -0.6218 | No | ||
| 57 | MNAT1 | 9396 2161 | 15363 | -0.569 | -0.6370 | No | ||
| 58 | PSMB6 | 9634 | 15579 | -0.698 | -0.6455 | No | ||
| 59 | PSMB4 | 15252 | 15677 | -0.765 | -0.6474 | No | ||
| 60 | PSMD4 | 15251 | 15753 | -0.824 | -0.6479 | Yes | ||
| 61 | POLR2A | 5394 | 15814 | -0.870 | -0.6474 | Yes | ||
| 62 | FEN1 | 8961 | 15855 | -0.898 | -0.6456 | Yes | ||
| 63 | GTF2F1 | 13599 | 15922 | -0.958 | -0.6450 | Yes | ||
| 64 | NUP62 | 9497 | 16051 | -1.023 | -0.6475 | Yes | ||
| 65 | TAF5 | 23833 5934 3759 | 16122 | -1.071 | -0.6466 | Yes | ||
| 66 | POLR2I | 12839 | 16178 | -1.117 | -0.6447 | Yes | ||
| 67 | PSMA5 | 6464 | 16241 | -1.174 | -0.6429 | Yes | ||
| 68 | PSMC6 | 7379 | 16310 | -1.237 | -0.6412 | Yes | ||
| 69 | RANBP2 | 20019 | 16336 | -1.259 | -0.6371 | Yes | ||
| 70 | PAK2 | 10310 5875 | 16358 | -1.281 | -0.6326 | Yes | ||
| 71 | CDK9 | 14619 | 16469 | -1.426 | -0.6323 | Yes | ||
| 72 | PSMD3 | 5803 | 16523 | -1.485 | -0.6287 | Yes | ||
| 73 | RNMT | 7501 | 16547 | -1.511 | -0.6234 | Yes | ||
| 74 | NUP155 | 2298 5027 | 16563 | -1.521 | -0.6176 | Yes | ||
| 75 | PSMD8 | 7166 | 16675 | -1.611 | -0.6166 | Yes | ||
| 76 | TBP | 671 1554 | 16689 | -1.625 | -0.6102 | Yes | ||
| 77 | RBX1 | 12130 7123 | 16729 | -1.662 | -0.6050 | Yes | ||
| 78 | KPNA1 | 22776 | 16790 | -1.735 | -0.6007 | Yes | ||
| 79 | PSMA4 | 11179 | 16871 | -1.842 | -0.5970 | Yes | ||
| 80 | SUPT4H1 | 9938 | 16949 | -1.956 | -0.5927 | Yes | ||
| 81 | PSMC4 | 2141 17915 | 16976 | -1.996 | -0.5854 | Yes | ||
| 82 | TAF10 | 10785 | 16990 | -2.022 | -0.5773 | Yes | ||
| 83 | TAF6 | 16322 891 | 17014 | -2.052 | -0.5696 | Yes | ||
| 84 | PSMC1 | 9635 | 17026 | -2.067 | -0.5611 | Yes | ||
| 85 | PSMB1 | 23118 | 17038 | -2.079 | -0.5527 | Yes | ||
| 86 | NUP37 | 3294 3326 19909 | 17158 | -2.232 | -0.5494 | Yes | ||
| 87 | POLR2E | 3325 19699 | 17221 | -2.334 | -0.5426 | Yes | ||
| 88 | TAF11 | 12733 | 17261 | -2.429 | -0.5341 | Yes | ||
| 89 | RDBP | 6589 | 17325 | -2.577 | -0.5263 | Yes | ||
| 90 | ERCC3 | 23605 | 17337 | -2.596 | -0.5155 | Yes | ||
| 91 | CCNH | 7322 | 17431 | -2.762 | -0.5085 | Yes | ||
| 92 | BANF1 | 6216 10706 3760 | 17446 | -2.797 | -0.4971 | Yes | ||
| 93 | TCEB1 | 13995 | 17465 | -2.829 | -0.4857 | Yes | ||
| 94 | PSMB3 | 11180 | 17536 | -2.970 | -0.4766 | Yes | ||
| 95 | GTF2E1 | 22597 | 17583 | -3.047 | -0.4658 | Yes | ||
| 96 | PSMD12 | 20621 | 17621 | -3.111 | -0.4542 | Yes | ||
| 97 | PSMD9 | 3461 7393 | 17635 | -3.147 | -0.4412 | Yes | ||
| 98 | PSMC2 | 16909 | 17677 | -3.270 | -0.4292 | Yes | ||
| 99 | PSMB5 | 9633 | 17739 | -3.411 | -0.4176 | Yes | ||
| 100 | NCBP2 | 12643 | 17747 | -3.425 | -0.4031 | Yes | ||
| 101 | GTF2E2 | 18635 | 17760 | -3.460 | -0.3886 | Yes | ||
| 102 | POLR2G | 23753 | 17842 | -3.679 | -0.3770 | Yes | ||
| 103 | NUP160 | 14957 | 17870 | -3.756 | -0.3621 | Yes | ||
| 104 | PSMA7 | 14318 | 17880 | -3.794 | -0.3460 | Yes | ||
| 105 | POLR2J | 16672 | 17905 | -3.892 | -0.3303 | Yes | ||
| 106 | TH1L | 14715 | 17931 | -3.986 | -0.3143 | Yes | ||
| 107 | PSMC5 | 1348 20624 | 17937 | -3.998 | -0.2972 | Yes | ||
| 108 | PSMD14 | 15005 | 17987 | -4.160 | -0.2817 | Yes | ||
| 109 | NUP107 | 8337 | 18011 | -4.236 | -0.2645 | Yes | ||
| 110 | POLR2H | 10888 | 18043 | -4.355 | -0.2472 | Yes | ||
| 111 | NUP93 | 7762 | 18055 | -4.422 | -0.2285 | Yes | ||
| 112 | GTF2H3 | 5551 | 18092 | -4.576 | -0.2105 | Yes | ||
| 113 | PSME3 | 20657 | 18155 | -4.800 | -0.1929 | Yes | ||
| 114 | RAE1 | 12395 | 18206 | -4.967 | -0.1740 | Yes | ||
| 115 | PSMD2 | 10137 5724 | 18238 | -5.062 | -0.1536 | Yes | ||
| 116 | KPNB1 | 20274 | 18273 | -5.185 | -0.1328 | Yes | ||
| 117 | GTF2H2 | 6236 | 18289 | -5.254 | -0.1107 | Yes | ||
| 118 | NUPL2 | 6072 | 18324 | -5.505 | -0.0886 | Yes | ||
| 119 | GTF2F2 | 21750 | 18491 | -6.869 | -0.0676 | Yes | ||
| 120 | NUP43 | 20094 | 18528 | -7.233 | -0.0380 | Yes | ||
| 121 | PSMD7 | 18752 3825 | 18607 | -9.809 | 0.0005 | Yes |