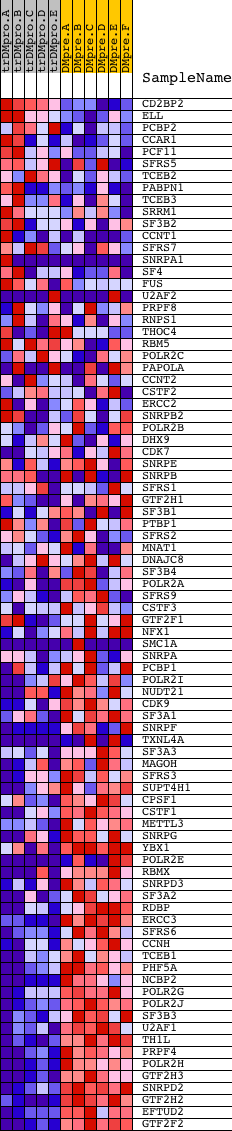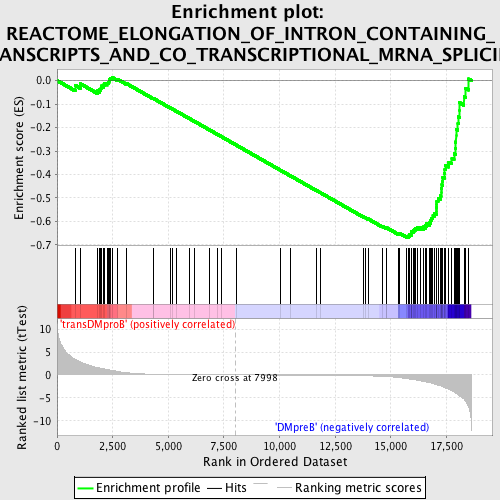
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | REACTOME_ELONGATION_OF_INTRON_CONTAINING_TRANSCRIPTS_AND_CO_TRANSCRIPTIONAL_MRNA_SPLICING |
| Enrichment Score (ES) | -0.6680001 |
| Normalized Enrichment Score (NES) | -1.6459875 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.027748983 |
| FWER p-Value | 0.342 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CD2BP2 | 17621 | 814 | 3.387 | -0.0207 | No | ||
| 2 | ELL | 18586 | 1041 | 2.850 | -0.0134 | No | ||
| 3 | PCBP2 | 9533 | 1814 | 1.620 | -0.0440 | No | ||
| 4 | CCAR1 | 7454 | 1917 | 1.496 | -0.0392 | No | ||
| 5 | PCF11 | 121 | 1970 | 1.442 | -0.0321 | No | ||
| 6 | SFRS5 | 9808 2062 | 1983 | 1.425 | -0.0230 | No | ||
| 7 | TCEB2 | 12563 23102 7476 | 2076 | 1.333 | -0.0188 | No | ||
| 8 | PABPN1 | 12018 | 2135 | 1.279 | -0.0132 | No | ||
| 9 | TCEB3 | 15707 | 2243 | 1.172 | -0.0110 | No | ||
| 10 | SRRM1 | 6958 | 2300 | 1.120 | -0.0063 | No | ||
| 11 | SF3B2 | 23974 | 2334 | 1.088 | -0.0006 | No | ||
| 12 | CCNT1 | 22140 11607 | 2349 | 1.081 | 0.0060 | No | ||
| 13 | SFRS7 | 22889 | 2401 | 1.027 | 0.0103 | No | ||
| 14 | SNRPA1 | 1373 18216 | 2484 | 0.986 | 0.0126 | No | ||
| 15 | SF4 | 18593 | 2716 | 0.771 | 0.0054 | No | ||
| 16 | FUS | 6135 | 3105 | 0.499 | -0.0121 | No | ||
| 17 | U2AF2 | 5813 5812 5814 | 4319 | 0.132 | -0.0766 | No | ||
| 18 | PRPF8 | 20780 1371 | 4324 | 0.131 | -0.0759 | No | ||
| 19 | RNPS1 | 9730 23361 | 5114 | 0.062 | -0.1181 | No | ||
| 20 | THOC4 | 5710 10114 | 5203 | 0.058 | -0.1224 | No | ||
| 21 | RBM5 | 13507 3016 | 5355 | 0.052 | -0.1302 | No | ||
| 22 | POLR2C | 9750 | 5936 | 0.034 | -0.1613 | No | ||
| 23 | PAPOLA | 5261 9585 | 6170 | 0.029 | -0.1737 | No | ||
| 24 | CCNT2 | 3993 13079 | 6832 | 0.016 | -0.2092 | No | ||
| 25 | CSTF2 | 24257 | 7209 | 0.011 | -0.2294 | No | ||
| 26 | ERCC2 | 1549 18363 3812 | 7384 | 0.008 | -0.2388 | No | ||
| 27 | SNRPB2 | 5470 | 8074 | -0.001 | -0.2760 | No | ||
| 28 | POLR2B | 16817 | 10054 | -0.030 | -0.3825 | No | ||
| 29 | DHX9 | 8845 4608 3948 | 10487 | -0.037 | -0.4056 | No | ||
| 30 | CDK7 | 21365 | 11650 | -0.061 | -0.4679 | No | ||
| 31 | SNRPE | 9843 | 11850 | -0.067 | -0.4782 | No | ||
| 32 | SNRPB | 9842 5469 2736 | 13755 | -0.181 | -0.5797 | No | ||
| 33 | SFRS1 | 8492 | 13847 | -0.190 | -0.5833 | No | ||
| 34 | GTF2H1 | 4069 18236 | 13978 | -0.204 | -0.5889 | No | ||
| 35 | SF3B1 | 13954 | 14620 | -0.312 | -0.6214 | No | ||
| 36 | PTBP1 | 5303 | 14798 | -0.355 | -0.6285 | No | ||
| 37 | SFRS2 | 9807 20136 | 14825 | -0.363 | -0.6274 | No | ||
| 38 | MNAT1 | 9396 2161 | 15363 | -0.569 | -0.6525 | No | ||
| 39 | DNAJC8 | 12712 | 15379 | -0.578 | -0.6493 | No | ||
| 40 | SF3B4 | 22269 | 15726 | -0.807 | -0.6625 | Yes | ||
| 41 | POLR2A | 5394 | 15814 | -0.870 | -0.6612 | Yes | ||
| 42 | SFRS9 | 16731 | 15840 | -0.887 | -0.6565 | Yes | ||
| 43 | CSTF3 | 10449 6002 | 15917 | -0.952 | -0.6541 | Yes | ||
| 44 | GTF2F1 | 13599 | 15922 | -0.958 | -0.6477 | Yes | ||
| 45 | NFX1 | 2475 | 15926 | -0.961 | -0.6413 | Yes | ||
| 46 | SMC1A | 2572 24225 6279 10780 | 16012 | -1.003 | -0.6390 | Yes | ||
| 47 | SNRPA | 7007 11992 7008 | 16045 | -1.019 | -0.6337 | Yes | ||
| 48 | PCBP1 | 17085 | 16099 | -1.053 | -0.6294 | Yes | ||
| 49 | POLR2I | 12839 | 16178 | -1.117 | -0.6259 | Yes | ||
| 50 | NUDT21 | 12665 | 16330 | -1.253 | -0.6255 | Yes | ||
| 51 | CDK9 | 14619 | 16469 | -1.426 | -0.6232 | Yes | ||
| 52 | SF3A1 | 7450 | 16568 | -1.527 | -0.6180 | Yes | ||
| 53 | SNRPF | 7645 | 16611 | -1.562 | -0.6096 | Yes | ||
| 54 | TXNL4A | 6567 11329 | 16753 | -1.689 | -0.6056 | Yes | ||
| 55 | SF3A3 | 16091 | 16797 | -1.744 | -0.5960 | Yes | ||
| 56 | MAGOH | 2386 16148 | 16830 | -1.789 | -0.5855 | Yes | ||
| 57 | SFRS3 | 5428 23312 | 16894 | -1.879 | -0.5760 | Yes | ||
| 58 | SUPT4H1 | 9938 | 16949 | -1.956 | -0.5655 | Yes | ||
| 59 | CPSF1 | 22241 | 17044 | -2.094 | -0.5562 | Yes | ||
| 60 | CSTF1 | 12515 | 17048 | -2.098 | -0.5420 | Yes | ||
| 61 | METTL3 | 21841 | 17063 | -2.118 | -0.5283 | Yes | ||
| 62 | SNRPG | 12622 | 17069 | -2.131 | -0.5139 | Yes | ||
| 63 | YBX1 | 5929 2389 | 17123 | -2.202 | -0.5017 | Yes | ||
| 64 | POLR2E | 3325 19699 | 17221 | -2.334 | -0.4909 | Yes | ||
| 65 | RBMX | 5367 9708 | 17257 | -2.417 | -0.4763 | Yes | ||
| 66 | SNRPD3 | 12514 | 17260 | -2.426 | -0.4598 | Yes | ||
| 67 | SF3A2 | 19938 | 17297 | -2.514 | -0.4445 | Yes | ||
| 68 | RDBP | 6589 | 17325 | -2.577 | -0.4283 | Yes | ||
| 69 | ERCC3 | 23605 | 17337 | -2.596 | -0.4111 | Yes | ||
| 70 | SFRS6 | 14751 | 17415 | -2.724 | -0.3966 | Yes | ||
| 71 | CCNH | 7322 | 17431 | -2.762 | -0.3785 | Yes | ||
| 72 | TCEB1 | 13995 | 17465 | -2.829 | -0.3609 | Yes | ||
| 73 | PHF5A | 22194 | 17601 | -3.077 | -0.3471 | Yes | ||
| 74 | NCBP2 | 12643 | 17747 | -3.425 | -0.3314 | Yes | ||
| 75 | POLR2G | 23753 | 17842 | -3.679 | -0.3113 | Yes | ||
| 76 | POLR2J | 16672 | 17905 | -3.892 | -0.2880 | Yes | ||
| 77 | SF3B3 | 18746 | 17915 | -3.934 | -0.2615 | Yes | ||
| 78 | U2AF1 | 8431 | 17929 | -3.980 | -0.2349 | Yes | ||
| 79 | TH1L | 14715 | 17931 | -3.986 | -0.2077 | Yes | ||
| 80 | PRPF4 | 7655 12845 | 18017 | -4.257 | -0.1831 | Yes | ||
| 81 | POLR2H | 10888 | 18043 | -4.355 | -0.1546 | Yes | ||
| 82 | GTF2H3 | 5551 | 18092 | -4.576 | -0.1259 | Yes | ||
| 83 | SNRPD2 | 8412 | 18097 | -4.591 | -0.0946 | Yes | ||
| 84 | GTF2H2 | 6236 | 18289 | -5.254 | -0.0689 | Yes | ||
| 85 | EFTUD2 | 1219 20203 | 18369 | -5.754 | -0.0338 | Yes | ||
| 86 | GTF2F2 | 21750 | 18491 | -6.869 | 0.0067 | Yes |