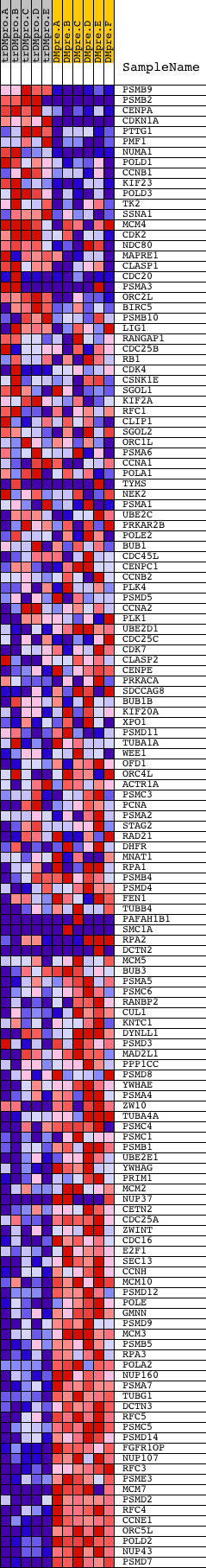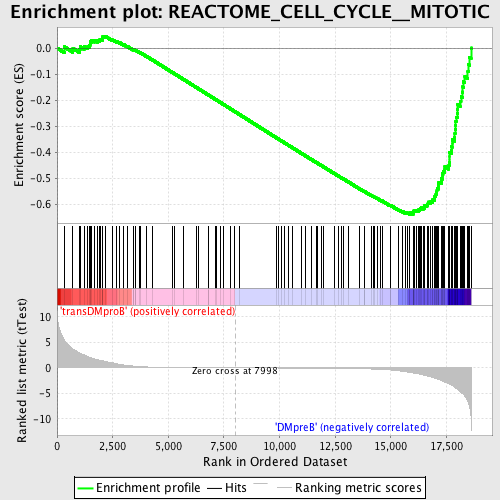
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | REACTOME_CELL_CYCLE__MITOTIC |
| Enrichment Score (ES) | -0.63988274 |
| Normalized Enrichment Score (NES) | -1.6717904 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.018728014 |
| FWER p-Value | 0.217 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMB9 | 23021 | 323 | 5.459 | 0.0050 | No | ||
| 2 | PSMB2 | 2324 16078 | 711 | 3.670 | -0.0008 | No | ||
| 3 | CENPA | 8736 | 1005 | 2.925 | -0.0046 | No | ||
| 4 | CDKN1A | 4511 8729 | 1034 | 2.865 | 0.0057 | No | ||
| 5 | PTTG1 | 20490 | 1212 | 2.512 | 0.0065 | No | ||
| 6 | PMF1 | 12452 | 1382 | 2.177 | 0.0063 | No | ||
| 7 | NUMA1 | 18167 | 1463 | 2.043 | 0.0104 | No | ||
| 8 | POLD1 | 17847 | 1481 | 2.020 | 0.0178 | No | ||
| 9 | CCNB1 | 11201 21362 | 1487 | 2.009 | 0.0258 | No | ||
| 10 | KIF23 | 19091 | 1549 | 1.916 | 0.0304 | No | ||
| 11 | POLD3 | 17742 | 1682 | 1.745 | 0.0305 | No | ||
| 12 | TK2 | 18779 | 1824 | 1.608 | 0.0295 | No | ||
| 13 | SSNA1 | 4 | 1905 | 1.514 | 0.0314 | No | ||
| 14 | MCM4 | 22655 1708 | 1971 | 1.440 | 0.0338 | No | ||
| 15 | CDK2 | 3438 3373 19592 3322 | 2032 | 1.374 | 0.0362 | No | ||
| 16 | NDC80 | 22900 | 2054 | 1.350 | 0.0407 | No | ||
| 17 | MAPRE1 | 4652 | 2057 | 1.347 | 0.0461 | No | ||
| 18 | CLASP1 | 14159 | 2154 | 1.262 | 0.0461 | No | ||
| 19 | CDC20 | 8421 8422 4227 | 2499 | 0.970 | 0.0315 | No | ||
| 20 | PSMA3 | 9632 5298 | 2673 | 0.808 | 0.0255 | No | ||
| 21 | ORC2L | 385 13949 | 2822 | 0.677 | 0.0203 | No | ||
| 22 | BIRC5 | 8611 8610 4399 | 3005 | 0.552 | 0.0127 | No | ||
| 23 | PSMB10 | 5299 18761 | 3162 | 0.463 | 0.0062 | No | ||
| 24 | LIG1 | 18388 1749 1493 | 3446 | 0.342 | -0.0078 | No | ||
| 25 | RANGAP1 | 2180 22195 | 3447 | 0.342 | -0.0063 | No | ||
| 26 | CDC25B | 14841 | 3504 | 0.319 | -0.0081 | No | ||
| 27 | RB1 | 21754 | 3542 | 0.305 | -0.0088 | No | ||
| 28 | CDK4 | 3424 19859 | 3699 | 0.256 | -0.0162 | No | ||
| 29 | CSNK1E | 6570 2211 11332 | 3766 | 0.239 | -0.0188 | No | ||
| 30 | SGOL1 | 7810 7809 13030 | 4020 | 0.178 | -0.0317 | No | ||
| 31 | KIF2A | 9217 11438 | 4284 | 0.137 | -0.0454 | No | ||
| 32 | RFC1 | 16527 | 5202 | 0.058 | -0.0948 | No | ||
| 33 | CLIP1 | 3603 7121 | 5270 | 0.056 | -0.0982 | No | ||
| 34 | SGOL2 | 14247 | 5692 | 0.041 | -0.1208 | No | ||
| 35 | ORC1L | 327 16144 | 6252 | 0.027 | -0.1510 | No | ||
| 36 | PSMA6 | 21270 | 6335 | 0.025 | -0.1553 | No | ||
| 37 | CCNA1 | 15340 | 6811 | 0.017 | -0.1810 | No | ||
| 38 | POLA1 | 24112 | 6819 | 0.017 | -0.1813 | No | ||
| 39 | TYMS | 5810 5809 3606 3598 | 7118 | 0.012 | -0.1973 | No | ||
| 40 | NEK2 | 5160 | 7143 | 0.012 | -0.1986 | No | ||
| 41 | PSMA1 | 1627 17669 | 7334 | 0.009 | -0.2088 | No | ||
| 42 | UBE2C | 12714 | 7495 | 0.007 | -0.2175 | No | ||
| 43 | PRKAR2B | 5288 2107 | 7813 | 0.002 | -0.2346 | No | ||
| 44 | POLE2 | 21053 | 7971 | 0.000 | -0.2431 | No | ||
| 45 | BUB1 | 8665 | 8213 | -0.003 | -0.2562 | No | ||
| 46 | CDC45L | 22642 1752 | 9842 | -0.026 | -0.3442 | No | ||
| 47 | CENPC1 | 8737 | 9942 | -0.028 | -0.3494 | No | ||
| 48 | CCNB2 | 19067 | 10074 | -0.030 | -0.3564 | No | ||
| 49 | PLK4 | 1870 1874 | 10201 | -0.032 | -0.3631 | No | ||
| 50 | PSMD5 | 2774 14612 | 10407 | -0.035 | -0.3740 | No | ||
| 51 | CCNA2 | 15357 | 10583 | -0.038 | -0.3833 | No | ||
| 52 | PLK1 | 9590 5266 | 10982 | -0.046 | -0.4047 | No | ||
| 53 | UBE2D1 | 19738 | 11180 | -0.050 | -0.4152 | No | ||
| 54 | CDC25C | 23468 1954 1977 | 11439 | -0.056 | -0.4289 | No | ||
| 55 | CDK7 | 21365 | 11650 | -0.061 | -0.4400 | No | ||
| 56 | CLASP2 | 13338 | 11689 | -0.062 | -0.4418 | No | ||
| 57 | CENPE | 15414 | 11897 | -0.069 | -0.4527 | No | ||
| 58 | PRKACA | 18549 3844 | 11953 | -0.071 | -0.4554 | No | ||
| 59 | SDCCAG8 | 14034 | 12474 | -0.092 | -0.4832 | No | ||
| 60 | BUB1B | 14908 | 12669 | -0.100 | -0.4933 | No | ||
| 61 | KIF20A | 23602 | 12795 | -0.107 | -0.4996 | No | ||
| 62 | XPO1 | 4172 | 12880 | -0.112 | -0.5037 | No | ||
| 63 | PSMD11 | 12772 7600 | 13078 | -0.123 | -0.5138 | No | ||
| 64 | TUBA1A | 1130 10233 5807 5806 | 13609 | -0.166 | -0.5418 | No | ||
| 65 | WEE1 | 18127 | 13796 | -0.184 | -0.5511 | No | ||
| 66 | OFD1 | 24007 | 13838 | -0.189 | -0.5526 | No | ||
| 67 | ORC4L | 11172 6460 | 14118 | -0.221 | -0.5668 | No | ||
| 68 | ACTR1A | 23653 | 14205 | -0.234 | -0.5705 | No | ||
| 69 | PSMC3 | 9636 | 14271 | -0.243 | -0.5730 | No | ||
| 70 | PCNA | 9535 | 14386 | -0.261 | -0.5781 | No | ||
| 71 | PSMA2 | 9631 | 14554 | -0.293 | -0.5859 | No | ||
| 72 | STAG2 | 5521 | 14608 | -0.308 | -0.5875 | No | ||
| 73 | RAD21 | 22298 | 14963 | -0.404 | -0.6050 | No | ||
| 74 | DHFR | 21590 | 15005 | -0.415 | -0.6055 | No | ||
| 75 | MNAT1 | 9396 2161 | 15363 | -0.569 | -0.6225 | No | ||
| 76 | RPA1 | 20349 | 15517 | -0.662 | -0.6280 | No | ||
| 77 | PSMB4 | 15252 | 15677 | -0.765 | -0.6335 | No | ||
| 78 | PSMD4 | 15251 | 15753 | -0.824 | -0.6341 | No | ||
| 79 | FEN1 | 8961 | 15855 | -0.898 | -0.6359 | Yes | ||
| 80 | TUBB4 | 22917 | 15857 | -0.900 | -0.6323 | Yes | ||
| 81 | PAFAH1B1 | 1340 5220 9524 | 15999 | -1.000 | -0.6358 | Yes | ||
| 82 | SMC1A | 2572 24225 6279 10780 | 16012 | -1.003 | -0.6323 | Yes | ||
| 83 | RPA2 | 2330 16057 | 16016 | -1.005 | -0.6283 | Yes | ||
| 84 | DCTN2 | 7635 12812 | 16040 | -1.017 | -0.6253 | Yes | ||
| 85 | MCM5 | 18564 | 16079 | -1.039 | -0.6231 | Yes | ||
| 86 | BUB3 | 18045 | 16170 | -1.110 | -0.6234 | Yes | ||
| 87 | PSMA5 | 6464 | 16241 | -1.174 | -0.6224 | Yes | ||
| 88 | PSMC6 | 7379 | 16310 | -1.237 | -0.6209 | Yes | ||
| 89 | RANBP2 | 20019 | 16336 | -1.259 | -0.6171 | Yes | ||
| 90 | CUL1 | 17462 | 16375 | -1.297 | -0.6138 | Yes | ||
| 91 | KNTC1 | 16705 | 16472 | -1.427 | -0.6131 | Yes | ||
| 92 | DYNLL1 | 12135 | 16518 | -1.483 | -0.6094 | Yes | ||
| 93 | PSMD3 | 5803 | 16523 | -1.485 | -0.6035 | Yes | ||
| 94 | MAD2L1 | 17422 | 16627 | -1.574 | -0.6026 | Yes | ||
| 95 | PPP1CC | 9609 5283 | 16670 | -1.602 | -0.5983 | Yes | ||
| 96 | PSMD8 | 7166 | 16675 | -1.611 | -0.5919 | Yes | ||
| 97 | YWHAE | 20776 | 16798 | -1.748 | -0.5913 | Yes | ||
| 98 | PSMA4 | 11179 | 16871 | -1.842 | -0.5876 | Yes | ||
| 99 | ZW10 | 3114 19464 | 16890 | -1.870 | -0.5808 | Yes | ||
| 100 | TUBA4A | 10232 | 16975 | -1.995 | -0.5771 | Yes | ||
| 101 | PSMC4 | 2141 17915 | 16976 | -1.996 | -0.5689 | Yes | ||
| 102 | PSMC1 | 9635 | 17026 | -2.067 | -0.5630 | Yes | ||
| 103 | PSMB1 | 23118 | 17038 | -2.079 | -0.5550 | Yes | ||
| 104 | UBE2E1 | 10243 5817 | 17073 | -2.141 | -0.5481 | Yes | ||
| 105 | YWHAG | 16339 | 17077 | -2.147 | -0.5394 | Yes | ||
| 106 | PRIM1 | 19847 | 17128 | -2.204 | -0.5330 | Yes | ||
| 107 | MCM2 | 17074 | 17133 | -2.214 | -0.5241 | Yes | ||
| 108 | NUP37 | 3294 3326 19909 | 17158 | -2.232 | -0.5162 | Yes | ||
| 109 | CETN2 | 24138 | 17268 | -2.458 | -0.5119 | Yes | ||
| 110 | CDC25A | 8721 | 17285 | -2.488 | -0.5025 | Yes | ||
| 111 | ZWINT | 19992 | 17308 | -2.540 | -0.4932 | Yes | ||
| 112 | CDC16 | 18676 | 17312 | -2.548 | -0.4829 | Yes | ||
| 113 | E2F1 | 14384 | 17346 | -2.611 | -0.4739 | Yes | ||
| 114 | SEC13 | 17039 | 17425 | -2.747 | -0.4668 | Yes | ||
| 115 | CCNH | 7322 | 17431 | -2.762 | -0.4557 | Yes | ||
| 116 | MCM10 | 14694 | 17609 | -3.085 | -0.4526 | Yes | ||
| 117 | PSMD12 | 20621 | 17621 | -3.111 | -0.4403 | Yes | ||
| 118 | POLE | 16755 | 17631 | -3.130 | -0.4279 | Yes | ||
| 119 | GMNN | 21513 | 17632 | -3.131 | -0.4150 | Yes | ||
| 120 | PSMD9 | 3461 7393 | 17635 | -3.147 | -0.4021 | Yes | ||
| 121 | MCM3 | 13991 | 17716 | -3.357 | -0.3926 | Yes | ||
| 122 | PSMB5 | 9633 | 17739 | -3.411 | -0.3797 | Yes | ||
| 123 | RPA3 | 12667 | 17754 | -3.445 | -0.3663 | Yes | ||
| 124 | POLA2 | 23988 | 17757 | -3.452 | -0.3522 | Yes | ||
| 125 | NUP160 | 14957 | 17870 | -3.756 | -0.3427 | Yes | ||
| 126 | PSMA7 | 14318 | 17880 | -3.794 | -0.3276 | Yes | ||
| 127 | TUBG1 | 20662 | 17890 | -3.827 | -0.3123 | Yes | ||
| 128 | DCTN3 | 15908 | 17911 | -3.923 | -0.2972 | Yes | ||
| 129 | RFC5 | 13005 7791 | 17920 | -3.951 | -0.2813 | Yes | ||
| 130 | PSMC5 | 1348 20624 | 17937 | -3.998 | -0.2657 | Yes | ||
| 131 | PSMD14 | 15005 | 17987 | -4.160 | -0.2512 | Yes | ||
| 132 | FGFR1OP | 23391 | 18000 | -4.196 | -0.2345 | Yes | ||
| 133 | NUP107 | 8337 | 18011 | -4.236 | -0.2176 | Yes | ||
| 134 | RFC3 | 12786 | 18116 | -4.657 | -0.2040 | Yes | ||
| 135 | PSME3 | 20657 | 18155 | -4.800 | -0.1863 | Yes | ||
| 136 | MCM7 | 9372 3568 | 18211 | -4.973 | -0.1688 | Yes | ||
| 137 | PSMD2 | 10137 5724 | 18238 | -5.062 | -0.1493 | Yes | ||
| 138 | RFC4 | 1735 22627 | 18260 | -5.128 | -0.1293 | Yes | ||
| 139 | CCNE1 | 17857 | 18329 | -5.540 | -0.1101 | Yes | ||
| 140 | ORC5L | 11173 3595 | 18464 | -6.552 | -0.0904 | Yes | ||
| 141 | POLD2 | 20537 | 18484 | -6.826 | -0.0632 | Yes | ||
| 142 | NUP43 | 20094 | 18528 | -7.233 | -0.0357 | Yes | ||
| 143 | PSMD7 | 18752 3825 | 18607 | -9.809 | 0.0005 | Yes |